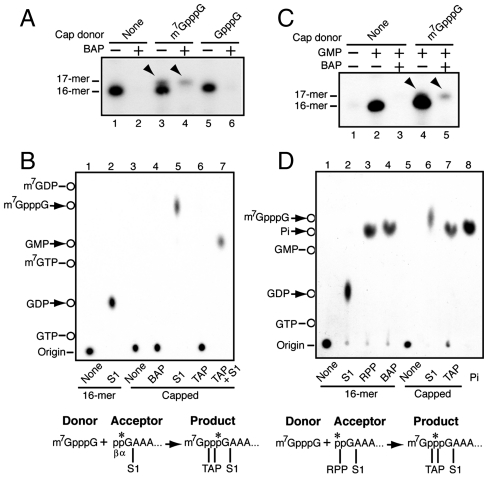
Fig. 2.
Preservation of 5′ diphosphate of L-A transcript in the triphosphate linkage of the capped product during cap snatching. (A) L-A virions were incubated in a CTP-omitted transcription mixture in the presence or absence of methylated or nonmethylated cap analogue (0.5 mM). Transcription was primed with [α-32P] GTP. BAP-treated or nontreated products were separated in 8 M urea/15% acrylamide gel. The arrowheads indicate BAP-resistant capped transcript (17-mer). (B) BAP-resistant 17-mer (capped) shown in A was isolated from the gel, treated with the enzymes singularly or sequentially as indicated, and analyzed by TLC using 0.3 M (NH4)2SO4 as solvent. Noncapped transcript (16-mer) was also processed in parallel as control. Deduced reaction scheme is drawn under the panel along with TAP and S1 cleavage sites. (C) L-A virions were incubated in a GTP- and CTP-omitted transcription mixture in the presence or absence of m7GpppG (0.5 mM). Transcription was primed with GMP in the presence of [γ-32P] ATP. BAP-treated or nontreated products were separated in an 8 M urea/15% acrylamide gel. The arrowheads indicate BAP-resistant capped transcripts (17-mer). (D) BAP-resistant 17-mer (capped) shown in C was isolated and processed as described in the legend to Fig. 1C except that TLC was carried out with 1 M LiCl as solvent. Noncapped transcript (16-mer) was also processed in parallel as control. RPP: RNA 5′ polyphosphatase that hydrolyses β- and γ- phosphates at the 5′ terminus of RNA. Lane 8: Pi standard. Deduced cap transfer reaction with some enzymatic cleavage sites is drawn under the panel. The asterisks show the positions of the 32P label.