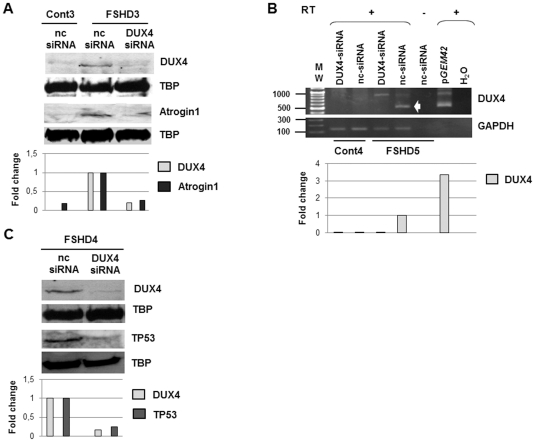
Figure 5. Evaluation of DUX4-siRNA efficiency on endogenous DUX4 and FSHD marker expression in FSHD primary myoblasts.
(A) 105 cells were seeded in 35 mm culture dish and directly transfected with negative control siRNA (nc-siRNA, 30 nM) or DUX4-siRNA3 (10 nM) using the reverse transfection method with 4 µl of siPORTNeoFX reagent. Differentiation was induced 4 hours after transfection, and cells were harvested 72 hours later. A nuclear extract was prepared and 20 µg of nuclear proteins were separated in parallel by two electrophoresis (12% PAGE-SDS), and transferred onto a nitrocellulose membrane. The proteins transfer was confirmed by Ponceau red staining. After rinsing the membranes were incubated either with 9A12 MAb or a polyclonal antibody against Atrogin1 (ECM Biosciences) followed by secondary antibodies coupled to horseradish peroxidase and revealed with the Femto Super Signal kit (Pierce). The antibodies were then stripped, and the same membranes revealed with an anti-TBP MAb (nuclear loading control). (B) Primary FSHD and control myoblasts transfected with the DUX4-siRNA (10 nM) or the negative control siRNA (nc-siRNA, 30 nM) were differentiated for 3 days. Total RNA was extracted. Reverse transcription was performed on 500 ng of DNase-treated total RNA with the 3′adaptator of the RLM-RACE kit (Ambion). 5 µl of the resulting cDNA were amplified by nested PCR (for details, see methods). The RT-PCR products were analysed by electrophoresis on an 1% agarose gel. A densitometry of the bands was performed for quantification. Data are normalized to GAPDH levels in each sample. pGEM42: expression vector containing 2 D4Z4 units (7); RT (+): with reverse transcriptase; (−): without reverse transcriptase. H2O: RT-PCR was performed with H2O. GAPDH: internal control. (C) Immunodetection of either DUX4 or TP53 with specific primary antibodies and appropriate secondary antibodies as described in the legend to Fig. 3 on two Western blots prepared with nuclear extracts of myotubes as described in Fig. 5A . A densitometry of the immunoreactive bands was performed. Data are normalized to TBP levels in each sample.