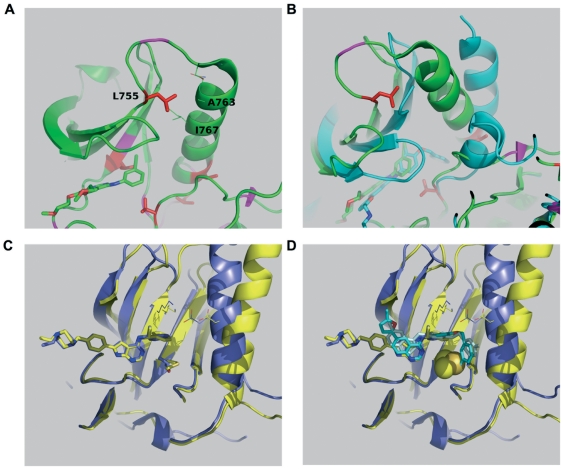
Figure 5. Structural analysis of lapatinib resistant ERBB2 kinase domain mutants.
(A) L755 packs against helix C, closest to residues Ala763 and Ile767, and makes no contacts with the inhibitors (structure 1M17 with inhibitor erlotinib is depicted lower left). (B) Comparing the active structure of 1M17 (green) to an inactive representative 1XKK bound to lapatinib shows the loss of L755 interactions (cyan). (C) Overlay of AEE788 bound structures of EGFR (2J6M, active, blue) and EGFR T790M (2JIU, inactive, yellow). The existence of the salt bridge linking the active site lysine K753 with the helix C E770 is a marker for the active state. The T798M (ERBB2 numbering) mutation does not significantly alter binding, although a rotation of the inhibitor aromat is apparent. (D) Superposition of two binding modes of lapatinib onto the overlay of figure 2C and display of the T798M atoms as Van der Waals spheres shows how the binding mode seen in 1XKK (cyan) obviously clashes with the mutation, but the binding mode of 3BBT (pale blue, ERBB4, which also has threonine as gatekeeper) does not.