Figure 5.
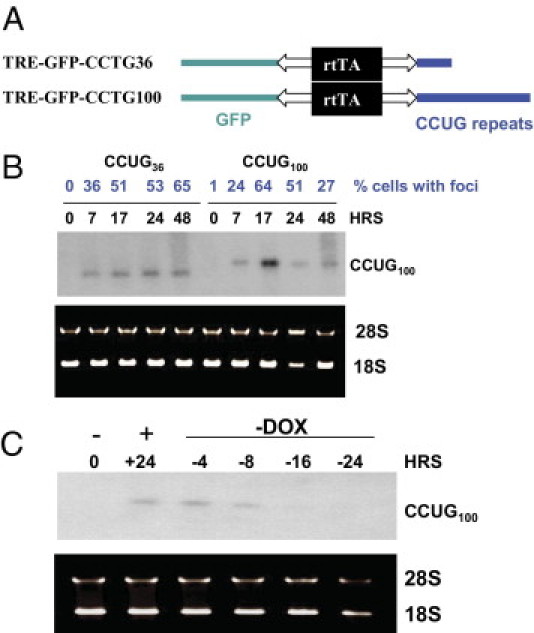
Regulation of transcription of CCUG36 and CCUG100 RNAs in monoclonal Tet-On cell cultures. A: Diagram shows the structure of TRE-GFP CCUG36 and CCUG100 constructs. B: Induction of short and long CCUG repeats after transcription pulse. Total RNA was collected from the double-stable CHO cells at 0, 7, 17, 24, and 48 hours after Dox addition and examined by Northern blot assay with 32P-CAGG10 probe. The number of cells with foci, described in Figure 6, are shown in blue on the top. RNA was stained with EtBr before transfer to nitrocellulose filter to evaluate quantity and quality of RNA. C: CCUG repeats degrade after Dox withdrawal. Northern blot analysis of CCUG100 RNA isolated from double-stable CHO cells after induction with Dox (+) and Dox withdrawal (−). Time points (in hours) at which RNA was extracted are shown on the top. Positions of RNA CCUG100 and 28S and 18S RNAs are also shown.
