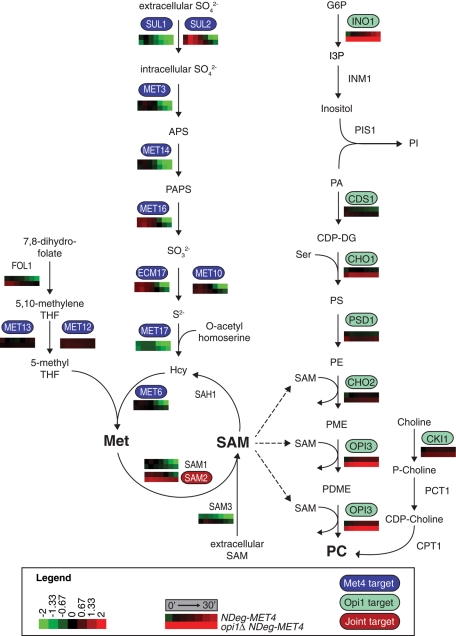
FIGURE 4:
Role of Met4p and Opi1p in regulating sulfur assimilation and PC biosynthesis pathways. Gene expression data for the biosynthesis genes were overlaid on the pathways. Gene expression time courses for a given gene are shown beneath that gene. Top and bottom, NDeg-MET4 and opi1Δ NDeg-MET4, respectively, with log2 fold change as indicated in the legend. Gene expression time courses are displayed as in the legend: from the instant before 1 μM estradiol treatment (0′) until 30 min after treatment (30′). Values at 0, 5, 15, and 30 min represent actual experimental measurements (indicated with black boxes above the time course), whereas values at 10, 20, and 25 min represent interpolated values (indicated with gray boxes above the time course) (see Materials and Methods). Genes directly regulated by Met4p (Lee et al., 2010) are shown in blue; genes directly regulated by Opi1p (Santiago and Mamoun, 2003) are shown in green; joint targets are shown in red. CDP-choline, cytidine diphosphate choline; CDP-DG, cytidine diphosphate diacylglycerol; APS, 5′-adenylylsulfate; G6P, glucose 6-phosphate; Hcy, homocysteine; I3P, inositol 3-phosphate; Met, methionine; P-choline, phosphocholine; PA, phosphatidic acid; PAPS, 3′-phospho-5′adenylylsulfate; PC, phosphatidylcholine; PDME, phosphatidyl-dimethylethanolamine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PME, phosphatidyl-monomethyl-ethanolamine; PS, phosphatidylserine; S2−, sulfide; SAM, S-adenosylmethionine; SO32−, sulfite; SO42−, sulfate; THF, tetrahydrofolate.