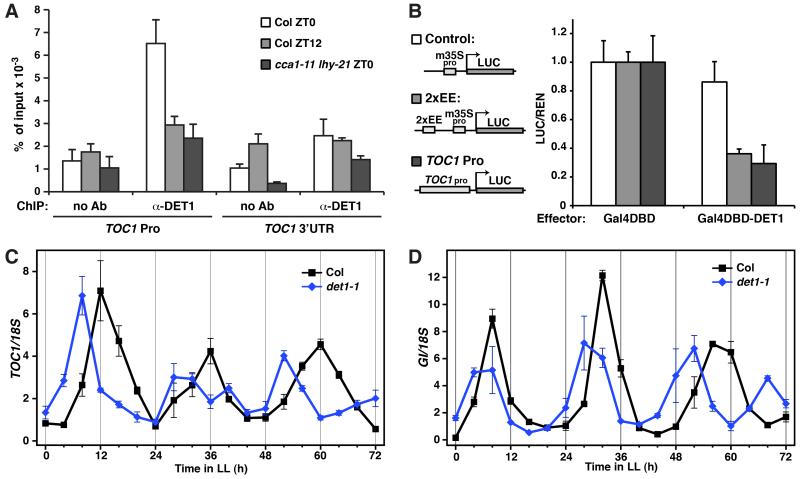
Figure 3. DET1 binds to target genes of CCA1 and LHY and regulates their circadian expressions.
(A) Chromatin Immunoprecipitation-quantitative PCR (ChIP-qPCR) analyses of DET1 binding on TOC1 promoter. ChIP assays were performed on seedlings of wild-type (Col) collected at zeitgeber time (ZT) 0 and 12 and the cca1-11 lhy-21 double mutant collected at ZT 0, with and without an anti-DET1 antibody. ChIP DNA was quantified by quantitative real-time PCR with primers and probes specific to the evening element (EE)-containing region of TOC1 promoter (TOC1 Pro) and the 3′ untranslated region of TOC1 (TOC1 3′UTR) (control). Signals were normalized to the input DNA (mean±s.d.; n≥3). Ab: Antibody (B) Transient transcriptional repression assays in plants for DET1 effect on an EE-containing reporter (2xEE) and a TOC1 promoter-driven reporter (TOC1 Pro). Assays were carried out as in Figure 1C. Relative reporter activities (LUC/REN) with the indicated effectors are shown (mean±s.d.; n≥4). Activities of reporters with the control effector, Gal4DBD, are set to 1. (C and D) Circadian rhythms of TOC1 (C) and GI (D) expression in wild-type (Col) and det1-1 under continuous light (LL). Plants were grown with long day cycles (16L8D) for 8 days and then released to LL. RNA levels were quantified by TaqMan real-time RT-PCR and signals were normalized to 18S (mean±s.d.; two biological and two technical replicates).