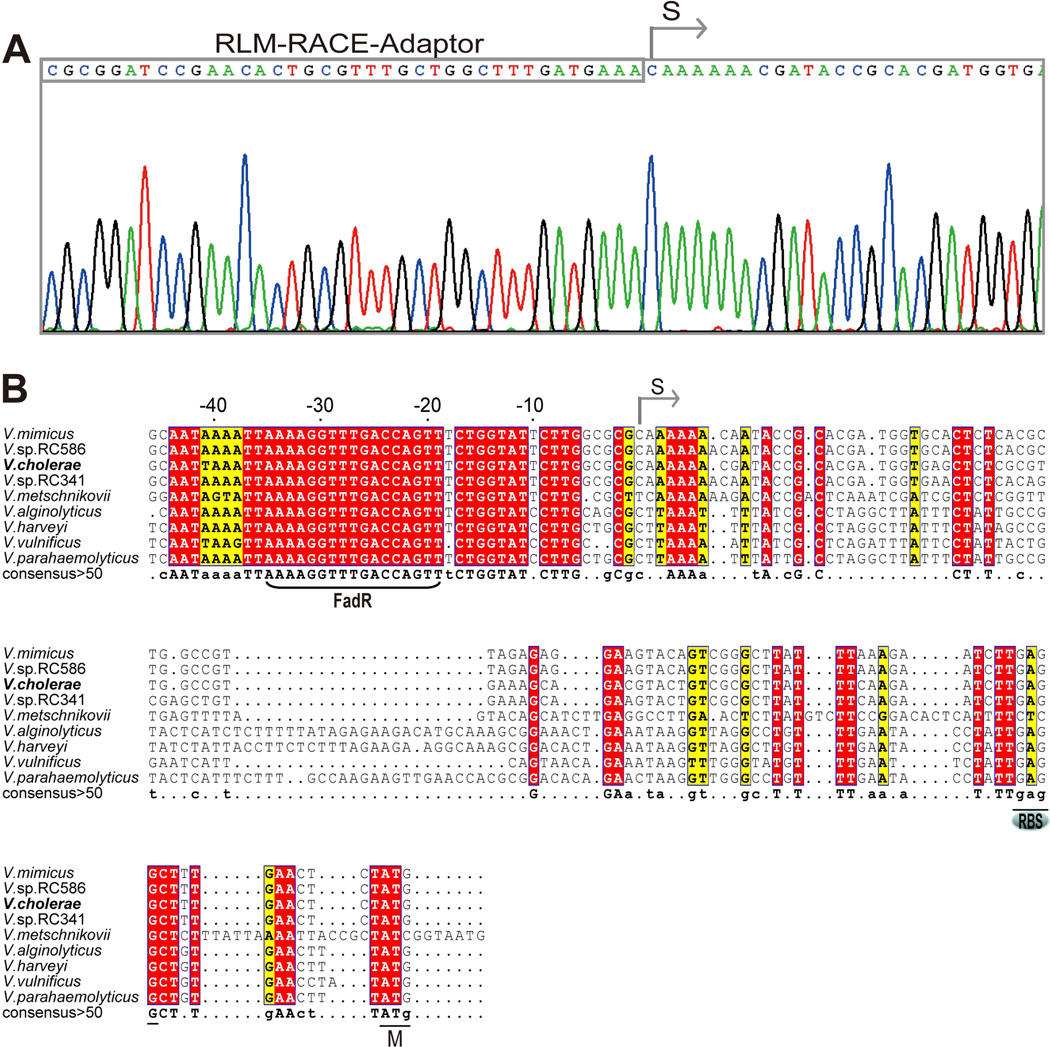
Fig. 4. Mapping of the V. cholerae plsB promoter.
A. 5’-RACE-based mapping of the V. cholerae plsB transcriptional start site
B. Multiple sequence alignments of V. cholerae plsB promoter region with those of some other Vibrio species. The predicted FadR-binding site is bracketed. Designations: S, transcriptional start site; RBS, ribosome binding site; M: initiator methionine. Identical residues are in given as white letters with red background, similar residues are in black letters with yellow background, varied residues are in grey letters and dots denote gaps. The plsB promoter region sequences are from V. mimicus (ACYU01000137.1, only one of four isolates with available contig sequences is given), V. vulnificus YJ016 (NC_005139.1), V. parahaemolyticus (BA000031), V. alginolyticus 40B (ACZB01000122.1), V. harveyi (CP000789), V. metschnikovii CIP (ACZO01000002.1), V. sp. RC586 (ADBD01000014.1), V. sp. RC341 (ACZT01000013.1), and V. cholerae ATCC14547 (results of direct sequencing in this study), respectively.