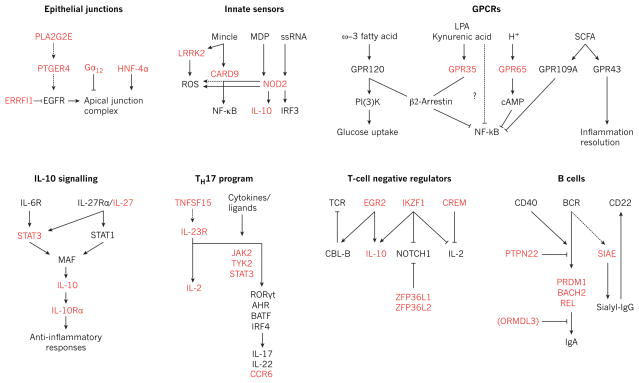
Figure 3. Genetic variants in IBD signalling modules.
Schematic of selected signalling pathways involved in the maintenance of intestinal homeostasis, including epithelial junctional complex assembly, innate immune recognition of pathogen-associated motifs, GPCRs and immune defence, anti-inflammatory interleukin-10 (IL-10) signalling, TH17-cell differentiation, inhibitory pathways in lymphocyte signalling, and B-cell activation and IgA antibody responses. Proteins encoded by genes identified as being in linkage disequilibrium with IBD-risk SNPs (r2 > 0.8) are highlighted in red. BCR, B-cell receptor; cAMP, cyclic AMP; EGFR, epidermal growth factor receptor; ERRFI1, ERBB receptor feedback inhibitor 1; Gα12, G protein subunit α12; GPCR, G-protein-coupled receptor; HNF-4α, hepatocyte nuclear factor-4α; LPA, lysophosphatidic acid; MDP, muramyl dipeptide; NF-κB, nuclear factor-κB; PI(3)K, phosphatidylinositol-3-OH kinase; PLA2G2E, phospholipase A2, group IIE; PTGER4, prostaglandin E receptor 4; SCFA, short-chain fatty acid; SIAE, sialic acid acetylesterase; ssRNA, single-stranded RNA; TCR, T-cell receptor.