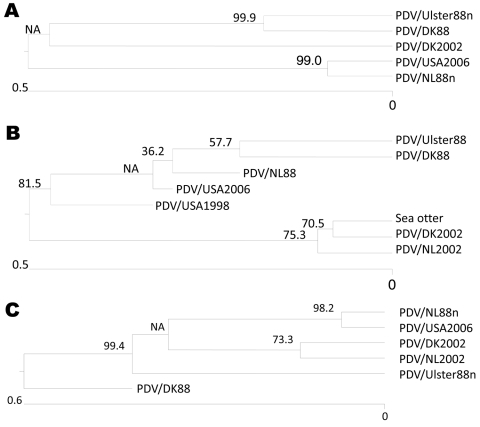
Figure 2.
Phylogenic relationship of PDV/USA2006 to viruses from the 1988 and 2002 epizootics in Europe based on hemagglutinin (H) and phosphoprotein (P) gene sequences. A) Phocine distemper virus (PDV) H gene sequences used for the alignments were from PDV/DK2002 (lung), GenBank accession no. FJ648456; PDV/DK88 (isolated in Vero cells), GenBank accession no. Z36979; PDV/Ulster88n (isolated in Vero cells), PDV/NL88n (blood), GenBank accession no. D10371 (minus described changes in this study); and PDV/USA2006 (isolated in VeroDogSLAM cells), GenBank accession no. 1375698. B) PDV P gene sequences used for the alignments were from PDV/DK2002 (lung), GenBank accession no. –af52587; PDV/NL2002 (lung) GenBank accession no. af52588; PDV/DK88 (isolated in Vero cells), GenBank accession no. –x75960; PDV/Ulster88 (isolated in Vero cells), GenBank accession no. D10371; PDV/NL88 (isolated in Vero cells), GenBank accession no. af525289; PDV/USA1998, GenBank accession no. ay3323389; and PDV/USA2006 (isolated in VeroDogSLAM cells), GenBank accession no. 1375683. Unrooted neighbor-joining phylogenetic trees in A and B were constructed by using the MegAlign version 7.1 package (DNASTAR, www.dnastar.com) with the ClustalW method (www.clustal.org). C) Unrooted neighbor-joining phylogenetic tree for concatenated H and P squences constructed by using ClustalV. Scale bars denote number of nucleotide substitutions per site along the branches. Percentage bootstrap values, indicating the significance of clusters, are shown.