Abstract
Interactions between proteins and their ligands play central roles in many physiological processes. The structural details for most of these interactions, however, have not yet been characterized experientially. Therefore, various computational tools have been developed to predict the location of binding sites and the amino acid residues interacting with ligands. In this manuscript, we assess the performance of 33 methods participating in the ligand binding site prediction category in CASP9. The overall accuracy of ligand binding site predictions in CASP9 appears rather high (average MCC of 0.62 for the ten top performing groups), and compared to previous experiments more groups performed equally well. However, this should be seen in context of a strong bias in the test data towards easy template based models. Overall, the top performing methods have converged to a similar approach using ligand binding site inference from related homologous structures, which limits their applicability for difficult “de novo” prediction targets. Here, we present the results of the CASP9 assessment of the ligand binding site category, discuss examples for successful and challenging prediction targets in CASP9, and finally suggest changes in the format of the experiment to overcome the current limitations of the assessment.
Keywords: protein function, protein structure, evaluation, assessment, binding site, active site, co-factor, ligand, CASP
Introduction
To perform their functions, proteins interact with a plethora of small molecules within the cell. Most of these interactions are unspecific and transient in nature (e.g. interactions with water and ions), some are persistent and may play a structural or functional role (e.g. certain metal ions), and others might be transient but nevertheless highly specific, often resulting in essential changes of the protein or the ligand (e.g. enzyme-substrate complexes or receptor-ligand complexes). Hence, the identification of a protein’s functionally important residues, such as ligand binding sites or catalytic active residues, is a crucial step towards the goal of understanding the protein’s molecular function and its biological role in the cell. Although protein ligand interactions are crucial for the function of a protein, in many cases they are unknown. While the kind of ligands interacting with a protein is often known from biochemical analyses, elucidating the structural details of these interactions requires elaborate and time-consuming studies by X-ray crystallography or NMR. Therefore, computational tools have been developed aiming at predicting the precise location of binding sites, and specifically which amino acid residues in a protein are directly interacting with ligands. Various approaches for the prediction of ligand binding sites have been proposed,1 both from structure and from sequence, based on sequence conservation 2–7, geometric criteria of the protein surface 8–12 or homology transfer from known structures.13–17
The function prediction category (FN) was introduced in the 6th Critical Assessment of Protein Structure Prediction (CASP), where predictions for Gene Ontology molecular function terms, Enzyme Commission numbers, and ligand binding site residues were evaluated. 18,19 Since very little new functional information becomes available during and after the experiment, the first two categories were difficult to assess. Therefore, since CASP8 the prediction task has been to identify functionally important residues such as ligand binding residues or catalytic residues. 20 Here, we present the assessment of 33 groups participating in the recent CASP9 experiment. In the ligand binding site prediction category (FN), the sequence of a protein with unknown structure was provided to predictors. The task was to predict the residues directly involved in ligand binding in the experimental control structure. This approach differs significantly from typical ligand binding studies (like docking or virtual screening), where the chemical identity of the ligand is given, and the correct geometric orientation of the molecule in the receptor protein is to be determined. 21–25 In CASP however, the chemical identity of the ligand is unknown at the time of prediction, and only the interacting residues are predicted.
In summary, all top performing groups have applied a similar approach, using ligand information derived from homologous structures in the PDB.26 In comparison to CASP8,20 we could not observe a significant progress by the top groups, but rather a larger number of groups performing at the same level. We believe that this observation is caused on one side by the bias in the data set to “easy” template based predictions with only a very small number of difficult “de novo” targets in recent rounds of CASP. This gives strong advantage to methods using PDB information directly, but discourages the development of methods addressing the more challenging “de novo” cases. Another limiting factor is the binary format of the prediction task, which does not allow specifying probabilities for specific residues or differentiating between types of ligands.
Materials and Methods
Prediction Targets
All CASP9 target structures were analyzed for non-solvent non-peptidic ligand groups in the deposited protein structures. Based on literature information, UniProt 27 annotations, structures of closely related homologues (Table SI, Supplementary Material), and conservation of functionally important residues, we aimed at identifying ligands with biological/functional relevance for the specific protein. All targets, including those containing ligands classified as “non-biologically relevant”, were further analyzed to indentify cases where a ligand clearly mimicked the interactions of known biologically relevant ligands for this target.
Binding Site Definition
For each prediction target, binding site residues were defined as those residues in direct contact with the ligand in the target structure, i.e. all protein residues with at least one heavy atom within a certain distance from any heavy atom of the ligand. The distance cutoff was defined by the CASP organizers as the sum of the van der Waals radii of the involved atoms plus a tolerance of 0.5 Å. In addition, different tolerance values ranging from 0 to 2.0 Å were evaluated.
In cases where multiple chains with bound ligands were present in the target structure (e.g. homo-oligomeric assemblies), the definition of the binding site residues for individual chains were combined into a single binding site definition. For targets where ligands were observed to bind in the interface between multiple chains, the oligomeric structure as defined by the authors and PISA 28 (5 cases) or only PISA (1 case) was used for the binding site definition. Analysis of structures and ligand binding sites were performed using OpenStructure (version 1.1). 29
For targets in which only part of the relevant ligand was present, the binding site definition was extended to include the entire biologically relevant ligand. In these cases, two separate evaluations of the prediction performance were conducted. First, denoted as ‘extended binding site’, all atoms of the partial and the extended ligand were used to define the binding site in the same way as described above. Second, denoted as ‘partial binding site’, only atoms of the partial ligand were used to define the binding site, whereas all residues exclusively in contact with the extended part of the ligand were treated as neutral and excluded from the evaluation.
Binding Site Prediction Evaluation
As in the previous assessment,20 binding site prediction performance was measured using the Matthews Correlation Coefficient30 (MCC) which accounts both for over and under predictions. For each target, residue predictions were classified as true positives (TP: correctly predicted binding site residues), true negatives (TN: correctly predicted non-binding site residues), false negatives (FN: incorrectly under predicted binding site residues), false positives (FP: incorrectly over predicted non-binding site residues) based on the binding site definition described before. The MCC was computed using Eq. 1:
MCC ranges from +1 (perfect prediction), over 0 (random prediction) to -1 (inverse prediction). Empty submissions which did not include any binding site predictions and missing predictions were assigned a MCC score of zero.
To reduce the effects of target difficulty on the ranking, MCC scores were standardized by computing Z scores among all predictions P for a given target T using Eq. 2:
In this equation, MCCP,T is the raw MCC score for target T given by predictor P, is the mean MCC score for target T, σT is the standard deviation of MCC scores for target T. The overall performance for each predictor was computed as the mean of Z scores over all targets, which was subsequently used for obtaining a final ranking. In addition to the MCC score, we computed the recently published binding site distance test (BDT) 31. BDT takes the actual three dimensional locations of the predicted residues into account and scores residues differently, according to the distance between the predicted and the observed binding site. Predictions close to the binding site score higher than more distant predictions. The BDT score ranges from 0, for a random prediction to 1, for a perfect prediction.
Robustness and significance
Statistical significance of the ranking and robustness with regard to composition of the target data set was assessed using two different methods. First, two-tailed Student’s paired t-tests as well as Wilcoxon signed rank tests32 between all predictor groups were performed based on MCC scores for each target. Both T-tests and Wilcoxon signed rank tests were performed using R (version 2.11.1). 33 Second, bootstrapping was performed, where scores were computed on a randomly selected subset of ¾ of all targets (i.e. 23 of 30 targets). 75 rounds of bootstrapping were executed for different target subsets, and for each bootstrapping experiment, mean, minimum and maximum Z scores per group were calculated as previously described. Additionally, the rank for each prediction group was calculated and mean, minimum and maximum ranks over all bootstrapping experiments were computed.
To assess the performance of groups on different types of ligands, we have analyzed the prediction performance separately on targets including only metal ions (10 targets) and on targets including only non-metal ligands (17 targets). Mixed targets including both metal and non-metal ligands (3 targets) were not considered in this sub-analysis.
Results and Discussion
Overall performance
In the CASP9 protein binding sites prediction category (FN), the predictors were given a protein sequence with unknown structure and asked to identify the residues involved in ligand binding. According to the CASP format, the predictions were binary and thus, classified each residue as either binding-site or non-binding-site residue. As defined by the organizers, only protein-small molecule interactions were considered in this category. The assessment of this category consisted of the following three steps: (1) identification of biologically relevant ligands in the target structures, (2) definition of binding site residues, (3) assessment of the prediction performance.
One dominant factor in assessing the correctness of ligand binding site prediction is the availability of experimental data, and the evaluation of the biological relevance of the specific ligand binding. Whether a certain ligand is observed in an experimental structure is first and foremost determined by the specific purification procedure, by the experimentalist’s choice of using this ligand for a co-crystallization experiment, and the specific experimental conditions (ligand concentration, pH and buffer conditions, ionic strength, precipitant etc.). If a ligand is not observed in a specific experimental structure, it could still bind under different conditions, i.e. it cannot be considered as a “true negative” data point for the assessment. On the other hand, if a certain ligand is observed in a target structure, we can classify the residues within this structure into “binding” and “non binding” with regard to this specific ligand. Note that a target protein might be able to bind different ligands under different experimental conditions, and only a subset of them might be present in the target structure at hand. For example, the structure of an enzyme might be crystallized in complex with the cofactor, but without substrate or product molecules.
Although the identification of ligands in CASP9 was based only on experimentally observed ligands, it was still not straightforward to categorize their biological relevance. Although in 73% of the target structures in CASP9 various ligands were present, most of them were not considered biologically relevant but rather as originating e.g. from solvent, crystallization precipitant, or buffers. For the assessment, however, we included only ligands which we considered to be biologically relevant. The decision on biological relevance was done by manual curation, primarily based on the type and location of the ligand, literature information, and UniProt27 annotations. In addition, information from structurally closely related homologues and conservation of functionally important residues was used to guide the selection process. Using this approach, 16 target structures with biologically relevant ligands were selected out of the 109 targets available in CASP9 for the assessment.
In addition, we have analyzed all remaining heteroatomic groups, if they occupied binding sites which mimicked the interactions of a known biologically relevant ligand for this protein. In these cases, we defined an “extended binding site” consisting of all residues in contact with the known biologically relevant ligand. We were careful to include only targets where the assignment was unambiguous, in order to avoid the inclusion of false binding site definitions. Using this approach, the number of target structures in the FN category was extended by 14, yielding a total of 30 targets in this category (Table I).
Table I.
Summary of CASP9 targets with bound ligands.
| Target | PDB | Partial Ligand | Extended Ligand | Chemical Class | Interface | CASP Category |
|---|---|---|---|---|---|---|
| T0515 | 3MT1 | SO4 | PLP, LYS | Non-metal | A-B | TBM |
| T0516 | 3NO6 | IMD | PF1 | Non-metal | TBM | |
| T0518 | 3NMB | NA | Metal | TBM | ||
| T0521 | 3MSE | CA, CA | Metal | TBM | ||
| T0524 | 3MWX | GOL | GAL | Non-metal | TBM | |
| T0526 | 3NRE | PEG | GLA | Non-metal | TBM | |
| T0529 | 3MWT | MN | Metal | TBM | ||
| T0539 | 2L0B | ZN, ZN | Metal | TBM | ||
| T0547 | 3NZP | PLP | PLP, LYS | Non-metal | A-B | TBM |
| T0548 | 3NNQ | ZN | Metal | TBM | ||
| T0565 | 3NPF | CSA | DGL, ALA | Non-metal | TBM | |
| T0570 | 3NO3 | MG, GOL | Metal, Non-metal | TBM | ||
| T0582 | 3O14 | ZN | Metal | TBM | ||
| T0584 | 3NF2 | SO4 | DST, IPR | Non-metal | TBM | |
| T0585 | 3NE8 | ZN | Metal | TBM | ||
| T0591 | 3NRA | LLP | Non-metal | A-B | TBM | |
| T0597 | 3NIE | ANP | Non-metal | TBM | ||
| T0599 | 3OS6 | SO4 | ISC | Non-metal | TBM | |
| T0604 | 3NLC | FAD | Non-metal | TBM/FM | ||
| T0607 | 3PFE | ZN | ZN, BES | Metal, Non-metal | TBM | |
| T0609 | 3OS7 | TLA | GAL | Non-metal | TBM | |
| T0613 | 3OBI | EDO | GAR, NHS | Non-metal | TBM | |
| T0615 | 3NQW | MN, SO4 | MN, GPX | Metal, Non-metal | TBM | |
| T0622 | 3NKL | SO4 | NAD | Non-metal | TBM | |
| T0625 | 3ORU | ZN | Metal | TBM | ||
| T0629 | 2XGF | FE, FE, FE, FE, FE, FE, FE | Metal | A-B-C | FM | |
| T0632 | 3NWZ | COA | Non-metal | A-B-C | TBM | |
| T0635 | 3N1U | CA | Metal | TBM | ||
| T0636 | 3P1T | TLA | HSA, PLP | Non-metal | A-B | TBM |
| T0641 | 3NYI | STE | Non-metal | TBM |
Within the selected targets, ten were found in complex with metal ions (Ca, Fe, Mg, Mn, Na, Zn), and further 17 targets in complex with non-metal ligands (Table I). The latter included amino acids and derivatives, nucleotides, sugars, fatty acids and others. Additionally, in three cases non-metal ligands were coordinated to metal ions (Mg, Mn, Zn). In most of the targets, the ligand binding site was located within a monomer, while for six targets the ligand was bound in the interface between multiple chains: T0515, T0547, T0591, T0636 (dimeric structures), T0629 (trimeric structure) and T0632 (tetrameric structure). The ligands were bound between all chains of the oligomeric structure, except for T0632 where the ligand is bound to only three of the four chains. Following the identification of biologically relevant ligands, the binding site residues for those targets were defined as those residues directly in contact with the ligand. Atoms were considered to be in contact if they were within a distance of the sum of their van der Waals radii plus a tolerance distance. The list of binding site residues used in the assessment for each target is provided in Table SI (Supplementary Material). The tolerance distance was defined as 0.5 Å by the CASP organizers. We tested the influence of different values for the tolerance distance of the binding site definition and their influence on the assessment of prediction performance. No significant differences in the overall prediction performances were observed for different tolerance distances (Fig. S1, Supplementary Material).
The majority of FN targets in CASP9 were classified as template based modeling targets (TBM), and only two targets were free modeling (FM) targets: (1) target T0629, where the ligand binding domain had no template structure (Fig. 8C), (2) target T0604, where the ligand was bound between two domains where one was a template based modeling (constituting 90% of the binding site residues) and one a free modeling domain (constituting 10% of the binding site residues). This strong bias in the data set has direct consequences for the assessment, as it is to be expected that template-based prediction methods will perform much better than “de novo” methods in this context.
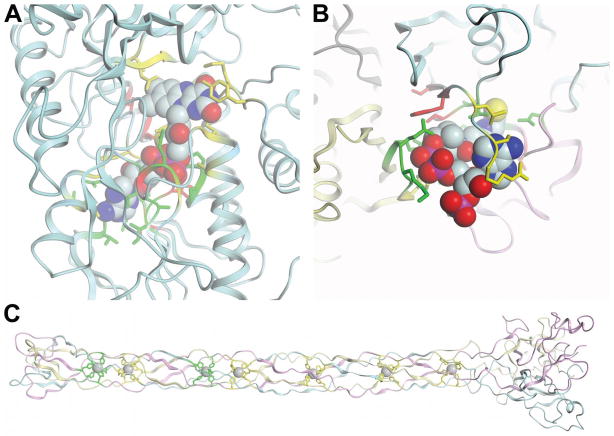
Fig. 8. Examples of binding site predictions.
All ligands are shown in spheres render mode. The protein backbone is shown in cartoon mode with each chain colored separately. All side chains of observed and predicted binding site residues are shown in licorice sticks. Correctly predicted residues (true positives) are colored in green, incorrectly under predicted binding site residues (false negatives) in yellow and incorrectly over predicted non-binding site residues (false positives) in red. (A) Target T0604 with predictions of group FN035. (B) Predictions of group FN096 for target T0632. (C) Group FN114’s predictions for target T0629.
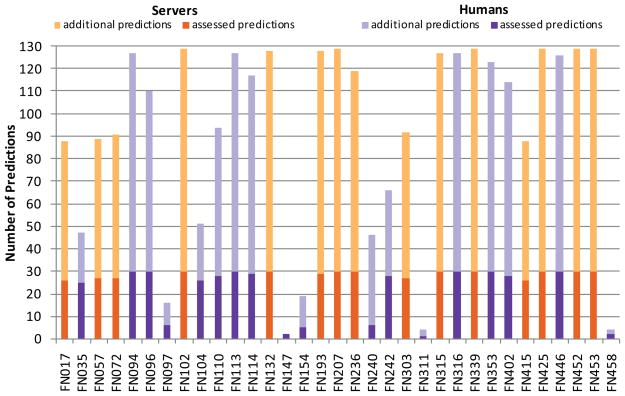
In total, 33 groups made predictions in the CASP9 FN category. A summary of the predictions is given in Fig. 1. Among the participating groups, 18 were registered as “human predictors” and 15 as “servers” (Table II). Most groups predicted at least 25 of the assessed 30 targets, i.e. 12 groups (6 humans, 6 servers) predicted between 25 and 29 of the assessed targets and 15 groups (6 humans, 9 servers) predicted all 30 targets; 6 human groups returned predictions for only 6 or less targets. Binding site prediction performance was measured using Z-scores of Matthews correlation coefficients (see Methods).a The comparison between all groups is shown in Fig. 2 where the error bars indicate minimum and maximum Z scores obtained by bootstrapping on a randomly selected subset of ¾ of the targets. The error bars indicate a fluctuation in the average Z score for each group. However, in case of a correlated movement in the score, this would not influence the groups ranking. Therefore, the rank for each prediction group was computed in each bootstrapping experiment and the average, minimum and maximum rank over all bootstrapping experiments is shown in Fig. 3.
Fig. 1. Overview of predictions per group.
Predictions for targets which were assessed in the FN category (i.e. targets with a relevant binding site) are displayed in dark colors, additional predictions which were not assessed (i.e. targets without an experimentally confirmed binding site) are displayed in light colors. Human groups are shown in purple, servers in orange.
Table II.
Groups participating in the FN category in CASP9.
| ID | Rank | Name | Type | Group |
|---|---|---|---|---|
| FN017 | 22 | 3DLIGANDSITE1 | S | Michael Sternberg |
| FN035 | 5 | CNIO-FIRESTAR | H | Gonzalo Lopez |
| FN057 | 21 | 3DLIGANDSITE3 | S | Michael Sternberg |
| FN072 | 23 | 3DLIGANDSITE4 | S | Michael Sternberg |
| FN094 | 8 | MCGUFFIN | H | Liam McGuffin |
| FN096 | 1 | ZHANG | H | Yang Zhang |
| FN097 | 30 | KOCHANCZYK | H | Marek Kochanczyk |
| FN102 | 15 | BILAB-ENABLE | S | Shugo Nakamura |
| FN104 | 7 | JONES-UCL | H | David Jones |
| FN110 | 6 | STERNBERG | H | Michael Sternberg |
| FN113 | 9 | FAMSSEC | H | Katsuichiro Komatsu |
| FN114 | 10 | LEE | H | Jooyoung Lee |
| FN132 | 27 | MN-FOLD | S | Chris Kauffman |
| FN147 | 28 | GENESILICO | H | Janusz Bujnicki |
| FN154 | 33 | JAMMING | H | Gabriel del Rio |
| FN193 | 24 | MASON | S | Huzefa Rangwala |
| FN207 | 26 | ATOME2_CBS | S | Jean-Luc Pons |
| FN236 | 12 | GWS | S | Jooyoung Lee |
| FN240 | 32 | TMD3D | H | Hiroshi Tanaka |
| FN242 | 4 | SEOK | H | Chaok Seok |
| FN303 | 20 | FINDSITE-DBDT | S | Jeffrey Skolnick |
| FN311 | 31 | ALADEGAP | H | Kei Yura |
| FN315 | 3 | FIRESTAR | S | Gonzalo Lopez |
| FN316 | 18 | LOVELL_GROUP | H | Simon Lovell |
| FN339 | 2 | I-TASSER_FUNCTION | S | Yang Zhang |
| FN353 | 17 | SAMUDRALA | H | Ram Samudrala |
| FN402 | 13 | TASSER | H | Jeffrey Skolnick |
| FN415 | 25 | 3DLIGANDSITE2 | S | Michael Sternberg |
| FN425 | 19 | INTFOLD-FN | S | Liam McGuffin |
| FN446 | 16 | KIHARALAB | H | Daisuke Kihara |
| FN452 | 11 | SEOK-SERVER | S | Chaok Seok |
| FN453 | 14 | HHPREDA | S | JohannesSoeding |
| FN458 | 29 | BILAB-SOLO | H | Mizuki Morita |
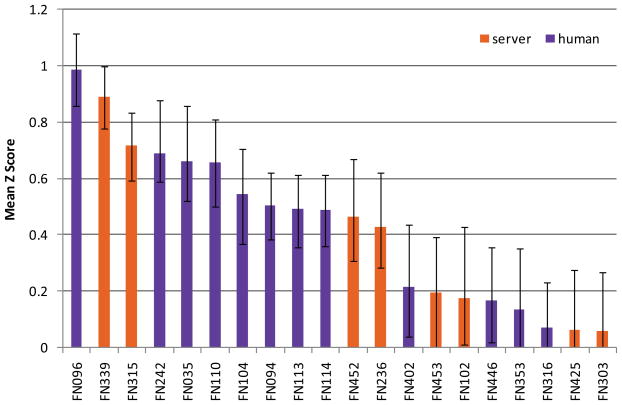
Fig. 2. Mean Z scores over all targets for the top 20 predictor groups.
Error bars show minimum and maximum average Z scores obtained from bootstrapping experiment. Human predictor groups are shown in purple, servers in orange.
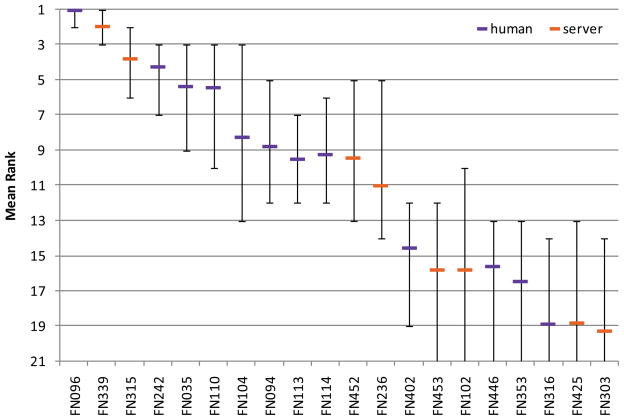
Fig. 3. Mean rank based on bootstrapping experiment for the top 20 predictor groups.
Error bars show minimum and maximum rank obtained from bootstrapping experiment. Human predictors are shown in purple, servers in orange.
The top 12 predictors clearly distinguished themselves from the following 21 groups and show a significantly better performance. Two predictors from the Zhang group (FN096, Zhang and FN339, I-TASSER_FUNCTION) show a better performance in terms of MCC compared to the following 10 groups, whereas the performance among those is comparable. Since many predictors seemed to perform similarly, statistical tests were used to assess the significance of the differences between these groups. Paired t-tests on all targets between all pairs of predictors were performed. The results are shown in Table III, with cells shaded according to computed P values. According to the t-test, the differences between the top ranked group (FN096, Zhang) and groups FN339 (I-TASSER_FUNCTION), FN242 (Seok) and FN035 (CNIO-Firestar) are not statically significant, while the differences between FN096 and the remaining predictors are significant. In addition, the non-parametric Wilcoxon signed rank test was performed, which yielded comparable results to the t-tests (Table SII, Supplementary Material). Recently, McGuffin and coworkers published an alternative binding site distance test (BDT) 31. Opposed to MCC, BDT takes the actual three dimensional positions of the predicted residues into account and scores residues differently, according to the distance between the predicted and the observed binding site. Hence, BDT limits the boundary effects originating from ambiguous definition of binding sites. When applying the BDT score on the predictions (Fig. S2, Supplementary Material), for the top ranked groups no significant deviations to the MCC based prediction assessment were observed. b
Table III. P-values computed by paired t-Test of all against all predictors.
Significant differences between two groups are indicated by cells with white background. For clarity, only the 12 top performing predictors are shown, sorted by their overall performance.
| FN096 | FN339 | FN315 | FN242 | FN035 | FN110 | FN104 | FN094 | FN113 | FN114 | FN452 | FN236 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FN096 | - | 0.24 | 0.01 | 0.08 | 0.06 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| FN339 | 0.24 | - | 0.27 | 0.20 | 0.28 | 0.20 | 0.05 | 0.04 | 0.02 | 0.05 | 0.02 | 0.02 |
| FN315 | 0.01 | 0.27 | - | 0.81 | 0.56 | 0.63 | 0.17 | 0.20 | 0.03 | 0.14 | 0.12 | 0.07 |
| FN242 | 0.08 | 0.20 | 0.81 | - | 0.85 | 0.90 | 0.31 | 0.28 | 0.27 | 0.19 | 0.10 | 0.09 |
| FN035 | 0.06 | 0.28 | 0.56 | 0.85 | - | 0.88 | 0.44 | 0.52 | 0.38 | 0.45 | 0.45 | 0.31 |
| FN110 | 0.01 | 0.20 | 0.63 | 0.90 | 0.88 | - | 0.33 | 0.28 | 0.27 | 0.30 | 0.33 | 0.18 |
| FN104 | 0.01 | 0.05 | 0.17 | 0.31 | 0.44 | 0.33 | - | 0.88 | 0.88 | 0.89 | 0.93 | 0.93 |
| FN094 | 0.00 | 0.04 | 0.20 | 0.28 | 0.52 | 0.28 | 0.88 | - | 0.99 | 0.98 | 0.94 | 0.79 |
| FN113 | 0.00 | 0.02 | 0.03 | 0.27 | 0.38 | 0.27 | 0.88 | 0.99 | - | 0.99 | 0.95 | 0.76 |
| FN114 | 0.00 | 0.05 | 0.14 | 0.19 | 0.45 | 0.30 | 0.89 | 0.98 | 0.99 | - | 0.96 | 0.56 |
| FN452 | 0.00 | 0.02 | 0.12 | 0.10 | 0.45 | 0.33 | 0.93 | 0.94 | 0.95 | 0.96 | - | 0.83 |
| FN236 | 0.00 | 0.02 | 0.07 | 0.09 | 0.31 | 0.18 | 0.93 | 0.79 | 0.76 | 0.56 | 0.83 | - |
As described above, for 14 targets the partial binding sites were individually extended around the observed ligand to reflect a binding site accommodating the most probable biologically relevant ligand. To investigate the influence of this extension, the assessment was performed both on all residues of the extended binding site and separately on all the residues of the partial binding site while treating the residues exclusively in the extended binding site as “neutral” for the analysis. For the top ranked groups no significant differences in the overall prediction performances were observed between partial and extended binding site definitions (Fig. S3, Supplementary Material). c
Assessment by type of binding sites
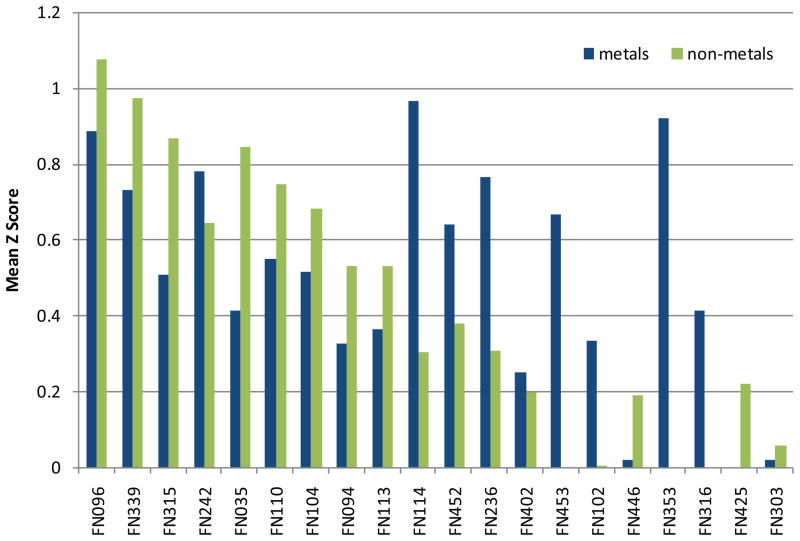
In addition to the overall performance, subsets of the targets were evaluated individually, according to the ligand’s chemotype. The distinct chemical properties of metal ions and organic ligands give raise to diverse binding sites. Thus, it could be expected that various prediction methods perform differently. To address this question, we have analyzed the prediction performance separately on all targets including only metal ligands (10 targets) and on targets including only non-metal ligands (17 targets). The mean Z-score per group separated into metal and non-metal targets are shown in Fig. 4. Within the top 10 groups most of them show a better performance for non-metal targets, with the exception of FN242 (Seok) and FN114 (Lee). Especially group FN114 shows a better performance on metal ligands, compared to an average performance on the full set of targets.
Fig. 4. Mean Z scores of the top 20 groups, separated by the ligand’s chemotype.
Metals are shown in blue, non-metals are shown in green.
Among the CASP9 FN targets, in six cases the ligand binds in the interface between multiple chains of an oligomeric protein complex. Although, the number of interface targets is very limited, we were interested in the question if the prediction of ligand binding sites of interface targets is more difficult than non-interface targets. We compared the average prediction performance, both according to mean MCC values, as well as the number of very good predictions (MCC > 0.85), for interface vs. non-interface targets. No significant difference was observed, thus on average, in those target categories it seems equally difficult to predict the binding site residues. However, it should be considered that four of the six targets are “trivial” oligomers, where a simple blast query returns a homologues template-ligand complex with the correct oligomeric state.
Human versus server predictions
Looking at the top 10 groups, 8 of them were registered as “humans”, and only 2 as “servers”. Overall, there is a striking difference between the average performance of human groups and server groups with a mean Z score of 0.47 and 0.15, respectively. Although predictor groups registered as “human” performed considerably better than “servers”, the role of human beings in the prediction process was difficult to evaluate. Several aspects seemed to contribute to this observation: Human predictors had access to multiple servers for structure modeling and various server binding site predictions, while server predictors have to rely on their own method only. While human predictors can make use of additional annotation from biological knowledgebases and scientific literature, servers have to rely on structured machine-readable information. A major bottleneck in this context seems the lack of consistent annotation of ligands found in PDB entries with respect to their biological relevance. It appears that human predictors benefit from the longer prediction time mainly by their ability to distinguish relevant from irrelevant ligand predictions.
Prediction methods have converged to a similar approach
When comparing the methods of the top performing groups, it seems they have converged to similar approaches, which are based on homology transfer from related structures in the PDB. By identifying homologous protein structures with bound ligands, putative binding site residues in the target model are classified by spatial proximity after alignment or superposition. The methods differ in their specific implementations with regards to the underlying structure databases (PDB vs. curated binding site libraries), target representation (alignment to structure vs. full atomic models), superposition to related structures to identify putative binding sites, and the use of residue conservation information in the prediction process. The major draw-back of these homology-based inference methods is that they rely on the availability of related protein structures with bound ligands and are thus unable to make predictions for novel proteins without prior ligand information.
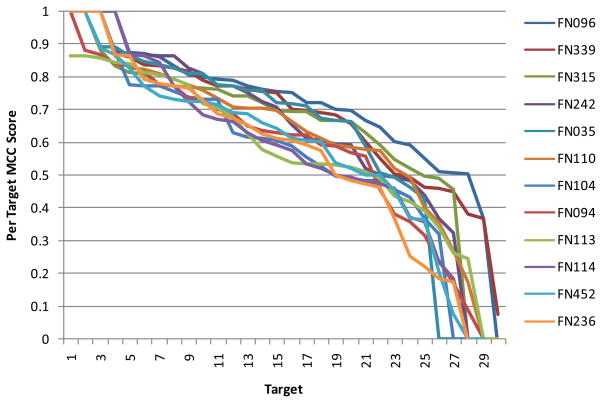
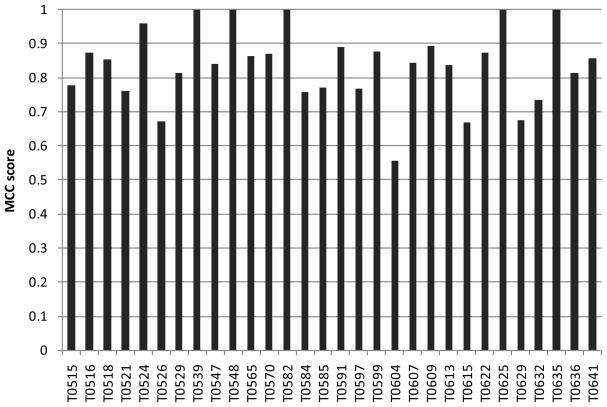
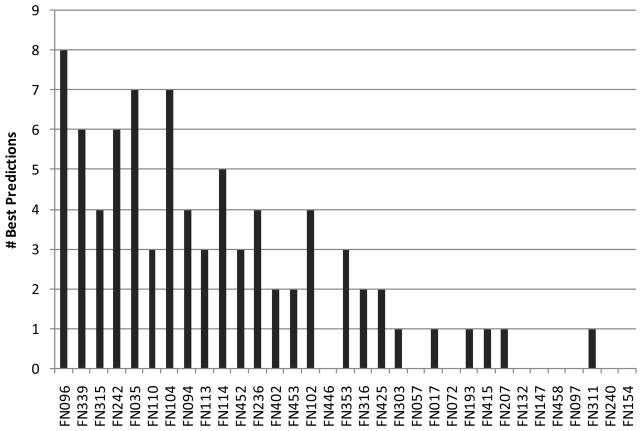
Although many groups have used similar approaches to make their predictions, we observed a surprising heterogeneity of performance within targets. As shown in Fig. 5 (and Fig. S4), the 12 top performing groups show overall a similar spectrum of results, with a few nearly perfectly predicted targets and some poorly predicted targets. Interestingly, when analyzing the results for individual targets, at least one good prediction was achieved across all groups (MCC value of at least 0.56; on average 0.84; see Fig. 6), and even predictors with a poor overall performance, can yield the best individual prediction for certain targets, as shown in Fig. 7. Thus, either the performance of the different methods is highly target specific, or there is a considerable random component in the prediction process in combination with a strong influence by the small and biased target data set.
Fig. 5. MCC scores for the 12 top performing groups for all targets.
Targets were sorted by their respective MCC score, individually for each group.
Fig. 6. Overall target difficulty.
MCC value of the best overall prediction for each target.
Fig. 7. Number of targets where a particular group returned the best prediction.
Groups are sorted by their overall performance. For one target, multiple groups can perform equally.
Prediction examples
Obviously, target T0604 was the most difficult target in the FN category in CASP9, with a maximum MCC score of 0.56 for the best prediction, and an average score of 0.29. The protein is a putative FAD dependent oxidoreductase with a bound FAD molecule (PDB:3nlc). The protein is monomeric and forms a large binding pocket for the ligand. The structure is shown in Fig. 8A together with the binding site predictions of group FN035 (CNIO-FIRESTAR) as one of the best predictions for this target. The top performing methods were able to accurately predict the lower part of the binding site around the adenine moiety, whereas all of them failed for the upper part of the binding site around the flavin moiety. This stems from the fact that this target structure has only remote homologues, which differ significantly in the flavin binding site region. This example clearly demonstrates the limitations of prediction methods that are based on homology transfer.
Target T0629 is the only target in the current ligand binding target set which was classified as free modeling target and thus has no template structure. The protein (PDB: 2xgf) is the bacteriophage T4 long tail fiber receptor-binding tip. It contains a long fiber like structure which is formed by three chains and binds seven iron atoms. Each iron atom is complexed by six histidine residues. Each protein chain contributes two histidines to each binding site, where the two histidines are in a His-X-His motive, with X being either Ser, Thr or Gly. The target structure is shown in Fig. 8C together with the binding site predictions of group FN114 (LEE), the best predictor for this target among the top 10. Common to all predictions for this target is that they correctly predicted a subset of the seven binding sites – most likely due to local similarity to another metal binding protein with a His-X-His motif, but no predictor identified all sites correctly.
The structure of target T0632 (PDB:3nwz) is a homo-tetramer which binds coenzyme-A. This ligand is interacting with three of the four chains of the protein, which seems to present a challenge for binding site residue prediction observed by a low average MCC of 0.22. An excellent prediction was obtained by group FN096 (Zhang) with an MCC of 0.72, which is depicted in Fig. 8B along with the target structure. Many residues were well predicted despite originating from different chains. In this prediction, the largest errors originate from missing some binding site residues due to an elongated terminus compared to structurally closely related templates.
Conclusion
The task of predicting binding sites from a protein’s sequence is of high relevance for life science research, ranging from functional characterization of novel proteins to applications in drug design, and consequently the ligand binding site prediction category in CASP has received increasing attention over the past years. In CASP9 it attracted a total of 33 predictors - ten more groups than in CASP8. In contrast to the previous CASPs, where only three predictors yielded reliable predictions,20 in this assessment nearly half of the prediction groups yielded reliable predictions for the majority of targets. Two groups (FN096, Zhang; FN339, I-TASSER_FUNCTION) performed better than the rest (when accounting for missing target predictions in the assessment), while the following ten prediction groups performed comparably well. This is not very surprising with respect to the observation that in this round all top performing groups based their methods on approaches, which are similar to the best performing strategy in previous CASP experiments (i.e. Sternberg34 and LEE15).
Limitations of the current format and recommendations for future experiments
The very low number of target structures with relevant ligands is a major limitation to the assessment as it does not allow to draw significant conclusions on the specific strengths and weakness of different prediction methods, e.g. with regard to target difficulty or type of the ligands. Only 30 of the total 109 CASP9 targets (28%) were considered to have a biologically relevant ligand bound in the target structures and were thus assessed in the FN category. It is likely that some of the remaining target proteins would bind interesting ligands under different experimental conditions, but such conclusions can not be made with the available data. In the previous CASP8 experiment, the total number of targets in this category was 27, illustrating that this is a recurring problem - and not specific to this round of CASP. Another rather drastic limitation of the FN category is the binary prediction format which classifies residues as either ligand binding/non-binding based on a hard distance cutoff. Consequently, all ligands are currently treated uniformly, independent of their chemical type, and all potential binding sites are treated uniformly, independent of their affinity (or binding probability) for different ligands. Moreover, most targets in the FN category were straightforward TBM targets with numerous, closely related template structures, and only one of the 30 targets was categorized as free modeling (FM). However, exactly this class of target structures is of highest interest for computational ligand binding site prediction, where no obvious information about the location of their binding sites is available. We would like to suggest the following modifications to the assessment of ligand binding site predictions to enable the community to benefit even further from future rounds of this experiment:
In order to accumulate a sufficiently large number of prediction targets, the assessment of this category should be done continuously based on a weekly PDB pre-release. This would allow assessing the performance in different ranges of target difficulty, similar to other CASP categories, and faciliate analyzing the strengths and weakness of different approaches. During the CASP meeting in Asilomar, we have suggested that the CAMEO project (Continuous Automated Model EvaluatiOn) of the Protein Model Portal 35 could contribute to this effort.
Binding sites differ chemically and structurally from each other e.g. a metal ion binding site has different characteristics compared to e.g. a sugar binding site. We therefore suggest that the assessment of binding site residue predictions should be made according to chemotype categories of the ligand expected to be bound. We would like to propose the following categories: “metal ions” (e.g. Na, Ca, Zn, Fe, Mn, Mg, etc.), “inorganic anions” (e.g. SO4, PO4), “DNA/RNA” for poly-ribonucleic acid binding sites, and “organic ligands” for cofactors, substrates and receptor agonists/antagonists (e.g. NAD, FAD, ATP, SAM, CoA, PLP, etc.). More fine grained assessment categories might be necessary if more specific prediction methods emerge in the future.
The binary prediction of binding site residues should be replaced by a continuous probability measure, thus reflecting the likelihood for a residue to be involved in binding a ligand of a certain type. For example a certain residue might be predicted as having a high probability to bind a metal ion, but a low probability to bind an organic ligand. The assessment of continuous prediction variable (e.g. using ROC type analysis) would better reflect the spectrum of “high affinity” and “low affinity” sites of different types.
The experimentalist solving a protein structure typically will have more insights and experimental evidence for the biological role and relevance of ligands observed in a protein structure than the information which is publicly available to assessors during the CASP experiment. It would therefore be beneficial to capture the information about the biological role of “HETATM” records during PDB deposition.
Predicting binding sites from a protein’s sequence has the potential for yielding high impact on life science research – if the predictions are specific and accurate enough to help addressing relevant biological questions. We hope that with the suggested modifications, the assessment of ligand binding site predictions will be more suited to evaluate the current state of the art of prediction methods, identify possible bottlenecks, and further stimulate the development of new methods.
Supplementary Material
Fig. S1 Influence of different binding site definitions on the prediction performance of the top 12 predictors. Mean Z score are shown for different tolerance distance used for binding site definition. All residues with a least one atom within the sum of the van der Waals radius plus the tolerance distance were considered to belong to the binding site.
Fig. S2 Comparison between the overall prediction performance evaluated using the Mathews Correlation Coefficient (MCC) and the Binding site Distance Test (BDT). Overall prediction performance is shown in mean Z Scores over all targets.
Fig. S3 Comparison of the overall prediction performance observed with the extended and the partial binding site definitions. Overall prediction performance is shown in mean Z Scores over all targets.
Fig. S4 MCC scores for all groups with at least 10 predictions for all targets. Targets were sorted by their respective MCC score, individually for each group.
Table SI Definition of binding site residues used in the assessment. [a] PDB id for the deposited target structure which was used to define the partial binding site. [b] PDB id of the protein structure related to the target structure, used in addition to define the extended binding site.
Acknowledgments
The authors would like to thank the experimental groups for providing the target structures for the CASP9 experiments, and all predictors for their participation. We are especially grateful to Mike Sternberg and Johannes Söding for fruitful discussions on ligand binding site prediction and assessment. This work was partially supported by the SIB Swiss Institute of Bioinformatics and by grant U01 GM093324-01 from the National Institute of General Medical Sciences.
Abbreviations
- MCC
Matthews’ Correlation Coefficient
- TBM
Template-Based Modelling
- FM
Free Modelling
Footnotes
As described in Materials and Methods, the authors decided that assigning a MCC score of zero to empty submissions which did not include any binding site predictions and to missing predictions would most appropriately reflect a “real life” prediction situation in the assessment. Please note that this policy has consequences for the final ranking as it penalizes methods which are not able to make predictions for some targets, and encourages the risky development of novel methods as there is no implicit penalty for making predictions for challenging targets.
The largest change in ranking by 3 positions would be for group FN110.
The largest difference was observed for group FN113 which would change rank by 3 positions.
References
- 1.Gherardini PF, Helmer-Citterich M. Structure-based function prediction: approaches and applications. Brief Funct Genomic Proteomic. 2008;7(4):291–302. doi: 10.1093/bfgp/eln030. [DOI] [PubMed] [Google Scholar]
- 2.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20(8):1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 3.Casari G, Sander C, Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995;2(2):171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- 4.del Sol A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol. 2003;326(4):1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- 5.Fischer JD, Mayer CE, Soding J. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24(5):613–620. doi: 10.1093/bioinformatics/btm626. [DOI] [PubMed] [Google Scholar]
- 6.Innis CA. siteFiNDER|3D: a web-based tool for predicting the location of functional sites in proteins. Nucleic Acids Res. 2007;35(Web Server issue):W489–494. doi: 10.1093/nar/gkm422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pazos F, Rausell A, Valencia A. Phylogeny-independent detection of functional residues. Bioinformatics. 2006;22(12):1440–1448. doi: 10.1093/bioinformatics/btl104. [DOI] [PubMed] [Google Scholar]
- 8.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62(2):479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 9.Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15(6):359–363. 389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez M, Ghersi D, Sanchez R. SITEHOUND-web: a server for ligand binding site identification in protein structures. Nucleic Acids Res. 2009;37(Web Server issue):W413–416. doi: 10.1093/nar/gkp281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13(5):323–330. 307–328. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 13.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105(1):129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez G, Valencia A, Tress ML. firestar - prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Research. 2007;35:W573–W577. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh M, Joo K, Lee J. Protein-binding site prediction based on three-dimensional protein modeling. Proteins-Structure Function and Bioinformatics. 2009;77:152–156. doi: 10.1002/prot.22572. [DOI] [PubMed] [Google Scholar]
- 16.Pandit SB, Brylinski M, Zhou H, Gao M, Arakaki AK, Skolnick J. PSiFR: an integrated resource for prediction of protein structure and function. Bioinformatics. 2010;26(5):687–688. doi: 10.1093/bioinformatics/btq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wass MN, Kelley LA, Sternberg MJE. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Research. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soro S, Tramontano A. The prediction of protein function at CASP6. Proteins. 2005;61 (Suppl 7):201–213. doi: 10.1002/prot.20738. [DOI] [PubMed] [Google Scholar]
- 19.Lopez G, Rojas A, Tress M, Valencia A. Assessment of predictions submitted for the CASP7 function prediction category. Proteins-Structure Function and Bioinformatics. 2007;69:165–174. doi: 10.1002/prot.21651. [DOI] [PubMed] [Google Scholar]
- 20.Lopez G, Ezkurdia I, Tress ML. Assessment of ligand binding residue predictions in CASP8. Proteins. 2009;77 (Suppl 9):138–146. doi: 10.1002/prot.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang SY, Grinter SZ, Zou X. Scoring functions and their evaluation methods for protein-ligand docking: recent advances and future directions. Phys Chem Chem Phys. 2010;12(40):12899–12908. doi: 10.1039/c0cp00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach AR, Shoichet BK, Peishoff CE. Prediction of protein-ligand interactions. Docking and scoring: successes and gaps. J Med Chem. 2006;49(20):5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 24.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 26.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35(Database issue):D301–303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Research. 2011;39(suppl 1):D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Biasini M, Mariani V, Haas J, Scheuber S, Schenk AD, Schwede T, Philippsen A. OpenStructure: a flexible software framework for computational structural biology. Bioinformatics. 2010;26:2626–2628. doi: 10.1093/bioinformatics/btq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 31.Roche DB, Tetchner SJ, McGuffin LJ. The binding site distance test score: a robust method for the assessment of predicted protein binding sites. Bioinformatics. 2010;26(22):2920–2921. doi: 10.1093/bioinformatics/btq543. [DOI] [PubMed] [Google Scholar]
- 32.Wilcoxon F. Individual Comparisons by Ranking Methods. Biometrics Bull. 1945;1(6):80–83. [Google Scholar]
- 33.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 34.Wass MN, Sternberg MJ. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins. 2009;77 (Suppl 9):147–151. doi: 10.1002/prot.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The Protein Model Portal. J Struct Funct Genomics. 2009;10(1):1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Influence of different binding site definitions on the prediction performance of the top 12 predictors. Mean Z score are shown for different tolerance distance used for binding site definition. All residues with a least one atom within the sum of the van der Waals radius plus the tolerance distance were considered to belong to the binding site.
Fig. S2 Comparison between the overall prediction performance evaluated using the Mathews Correlation Coefficient (MCC) and the Binding site Distance Test (BDT). Overall prediction performance is shown in mean Z Scores over all targets.
Fig. S3 Comparison of the overall prediction performance observed with the extended and the partial binding site definitions. Overall prediction performance is shown in mean Z Scores over all targets.
Fig. S4 MCC scores for all groups with at least 10 predictions for all targets. Targets were sorted by their respective MCC score, individually for each group.
Table SI Definition of binding site residues used in the assessment. [a] PDB id for the deposited target structure which was used to define the partial binding site. [b] PDB id of the protein structure related to the target structure, used in addition to define the extended binding site.