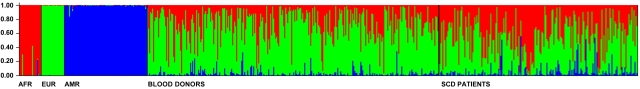
Figure 1.
Individual admixture in healthy blood donors and Sickle Cell Disease patients from Minas Gerais. Admixture was estimated using the method by Pritchard et al implemented in the software Structure.11 Each vertical bar represents an individual and his/her admixture proportions based on the parental populations on the left. Additional methodologic information and results are available as supplemental Methods. Structure was run using the following conditions and parameters: K = 3 (number of parental populations), burn-in period = 100 000, MCMC cycles after burn-in = 100 000, we used a priori information for the individuals from parental populations to assist the clustering (USEPOPINFO = 1), model = ADMIXTURE for the admixed individuals, α parameter was inferred for each population, GENSBACK = 2, MIGRPRIOR = 0.05, allele frequencies was assumed to be correlated.