Figure 3. NMN ameliorates hepatic insulin resistance and restores gene expression related to oxidative stress, inflammatory response, and circadian rhythm.
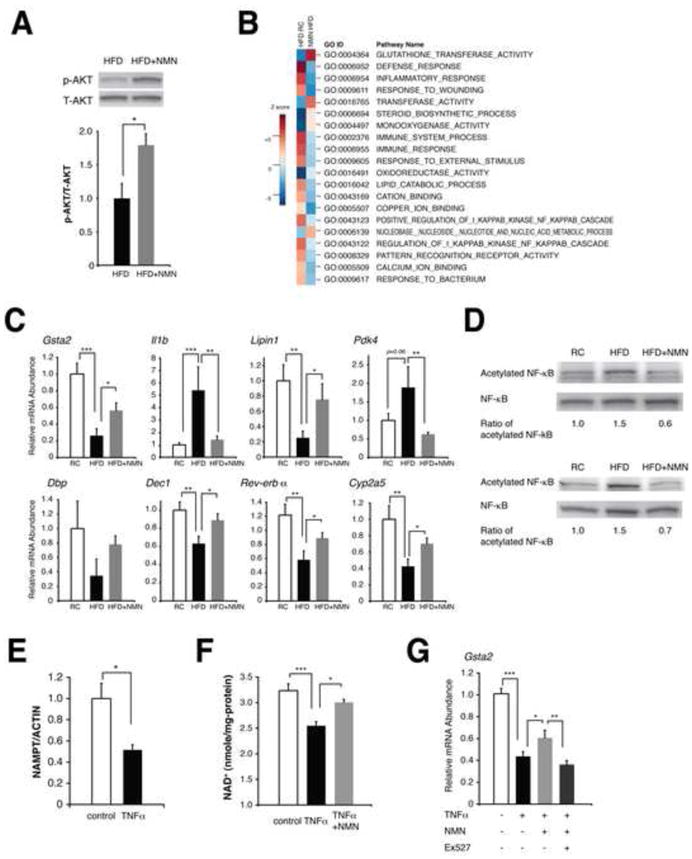
(A) The phosphorylation status of AKT in HFD and NMN-treated HFD female livers (n=3 mice per group). Signal levels of phosphorylated AKT were normalized to total AKT protein levels. (B) Biological pathways that were altered by HFD and reversed by NMN in female livers. Parametric analysis of gene-set enrichment (PAGE) was performed to identify pathways that were significantly up-regulated (red) or down-regulated (blue) by either HFD or NMN using our microarray data (n=4 mice for each condition). Twenty top pathways are listed following the sum of the absolute values of Z scores between two comparisons. (C) Quantitative RT-PCR results for representative genes related to oxidative stress, inflammatory response, circadian rhythm, and metabolism (n=4 to 5 mice per group). Gsta2, glutathione S-transferase alpha 2; Il1b, interleukin 1 beta; Pdk4, pyruvate dehydrogenase kinase isozyme 4; Dbp, D site of albumin promoter (albumin D-box) binding protein; Dec1, deleted in esophageal cancer 1. (D) Acetylation status of NF-κB p65 in RC, HFD, and NMN-treated HFD livers. Two independent sets of mice were used for this analysis, and numbers below each panel represent normalized ratios of acetylated to total p65 levels. (E–G) The effects of TNF-α on NAMPT-mediated NAD+ biosynthesis and gene expression in mouse primary hepatocytes. Cells were treated with 50 ng/ml TNF-α for 72 hrs and given indicated reagents for 6 hrs prior to harvesting for measurements. (E) NAMPT protein levels were normalized to ACTIN (n=3 per group). (F) TNF-α-treated cells were given 100 μM NMN prior to NAD+ measurements (n=3–6 per group). (G) TNF-α-treated cells were cultured with NMN or NMN plus 40 μM EX527 and examined for Gsta2 expression (n=6–9 per group). Data were analyzed by Student’s unpaired t test (A, E). Differences in Ct values or NAD+ levels were analyzed with one-way ANOVA with the Fisher’s PLSD post-hoc test (C, F, G). All values are presented as mean ± SEM. *P < 0.05; **P <0.01; ***P < 0.001.
