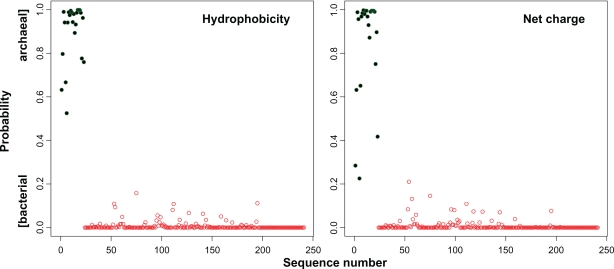
Figure 5.
A random forest (RF) can be trained to discriminate between bacterial and archaeal single-domain parvulins. Two descriptors were used (hydrophobicity and net charge) to describe the protein sequences. The very same dataset used for the MLP tree was used here. Prediction scale represents “0.0”, bacterial, and “1.0”, archaeal. The separation according to the MLP tree is represented in green (archaeal) and red (bacterial). For the hydrophobicity descriptor, the RF perfectly separate the two classes (F1 score = 1.0). For the net charge descriptor, the RF reaches an F1 score of 0.979 (cut off = 0.2).