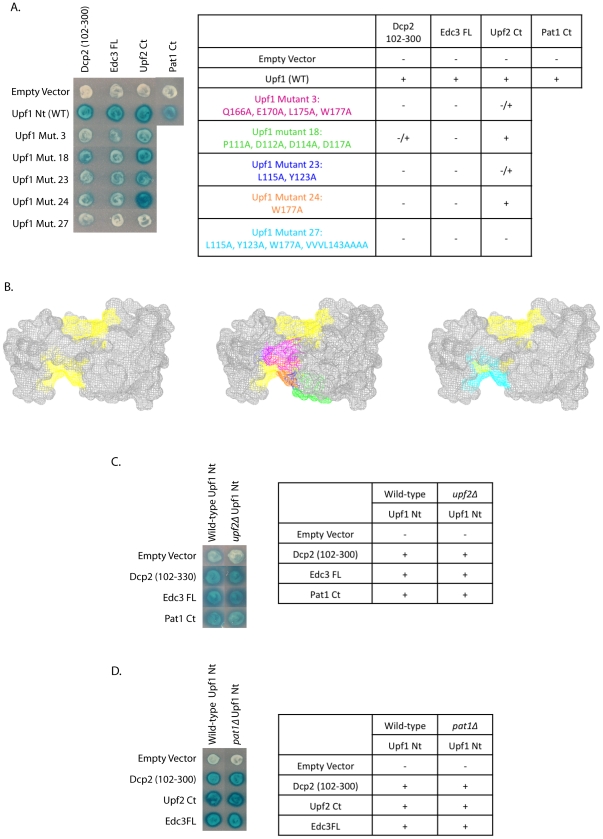
Figure 2. Edc3, Upf2 and Pat1 all interact with the N-terminal domain of Upf1; Edc3 and Upf2 do so in an overlapping, but not identical manner.
(A and B) Dcp2 (102–300), Edc3 FL, Upf2 Ct, and Pat1 Ct were all assessed for their ability to interact with Upf1 Nt (WT) by yeast two-hybrid analysis. Interactions between Upf1 Nt and Dcp2 (102–300), Edc3 FL, and Upf2 Ct were further characterized at the amino acid level. The S. cerevisiae Upf1 Nt structure was predicted using the human Upf1 crystal structure (PDB:2IYK) [13] and point mutations were made along the surface of Upf1. The predicted Upf2-binding sites are highlighted in yellow. Mutants were tested for interaction with Dcp2 (102–300), Edc3 FL, and Upf2 Ct by yeast two-hybrid analysis. Mutants which showed impaired interaction with Dcp2 (102–300), Edc3 FL, and/or Upf2 Ct are shown in pink, green, dark blue, orange, and light blue colors. (C) A upf2Δ strain was constructed and interaction between Upf1 Nt and Dcp2 (102–300), Edc3 FL and Pat1 Ct was assessed by yeast two-hybrid analysis. (D) A pat1Δ strain was constructed and interaction between Upf1 Nt and Dcp2 (102–300), Upf2 Ct, and Edc3 FL was assessed by yeast two-hybrid analysis.