Table 8.
Marine natural product structural revisions of multiple stereocenters (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Calafianin96(2000) Sponge |
NOE |

|
Total Synthesis97, 98 |
 Symbiodinolide99 (2007) Dinoflagellate |
Degradation NOE J-based |

|
Degradation Partial Synthesis Comparison100 |
 Clavosolide A101 (2002) Sponge |
J-based ROESY |

|
Total Synthesis102 |
 Laurentristich-4-ol103 (2005) Red alga |
NOE |

|
Total Synthesis104 |
 Vannusal A105 (1999) Ciliate |
NOE J-based |

|
Total Synthesis106, 107 |
 Vannusal B105 (1999) Ciliate |
NOE J-based Comparison |

|
Total Synthesis106, 107 |
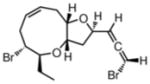
 Itomanallene A108(2002) Red alga |
NOE |
|
Total Synthesis109 |
 Asperdimin110 (2004) Fungus |
Comparison |

|
Total Synthesis111 |
 Aplysiallene112 (1985) Sea Hare |
OR NOE Comparison |

|
Total Synthesis113 |
 Aplysiol B114 (2010) Red alga |
NOE J-based Mol. Mod. |

|
Biogenesis NOE115 |
 Azaspiracid-1116(1998) Mussel |
NOE J-based |

|
Total Synthesis117 |
 Azaspiracid-2118 (1999) Mussel |
NOE Comparison |

|
Total Synthesis119 |
 Azaspiracid-3118(1999) Mussel |
NOE Comparison |

|
Total Synthesis119 |
 Amphidinolide H2120 (2002) Dinoflagellate |
NOE |

|
Total Synthesis121 |
 Hyrtiosterol122(2004) Coral |
NOE |

|
NOE123 |
 Pericosine A124 (1997) Fungus |
NOE J-based |

|
Total Synthesis125 |
 Callipeltoside C126 (1996) Sponge |
ROESY NOE |

|
Total Synthesis127 |
 Palmerolide A130 (2006) Tunicate |
Mosher J-based NOE |

|
Total Synthesis131, 132 |
 Neopeltolide133(2007) Sponge |
NOE |

|
Total Synthesis134 |
 Agardhilactone204(1996) Red alga |
Derivatization NMR |

|
Total Synthesis205 |
 Palau’amine206 (1993) Sponge |
ROESY J-based |

|
ROESY Mol. Mod. J-based207 |
 Bromopalau’amine208(1998) Sponge |
Comparison |

|
Comparison209 |
 Dibromopalau’amine208(1998) Sponge |
Comparison |

|
ROESY209 |
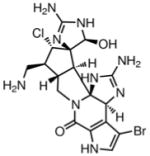
 Bromostyloguanidine208(1998) Sponge |
Comparison |
|
Comparison209 |
 Dibromostyloguanidine208(1998) Sponge |
Comparison |

|
Comparison209 |
|
Malyngamide U210 (2003) Cyanobacteria |
J-based NOESY |
|
Total Synthesis211 |
 Lucentamycin A212 (2007) Bacterium |
Marfey’s |

|
Total Synthesis213 |
 Nakiterpiosin214 (2004) Sponge |
NOE |

|
Total Synthesis215 |
 Kulokekahilide-2216 (2004) Mollusk |
Marfey’s |

|
Total Synthesis217 |
 Muurolane-derivative218(1997) Bacterium |
NOE J-based |

|
X-ray219 |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE219 |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE219 |
 Dolastatin 19221(2004) Mollusk |
NOE |

|
Total Synthesis222 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Predict. = predictions based on molecular modeling. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
