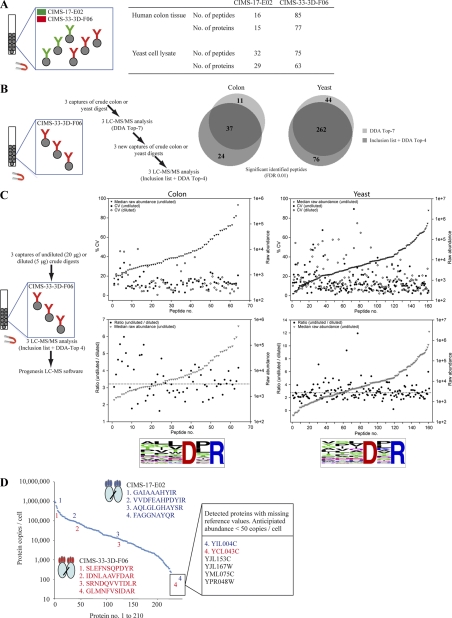
Fig. 3.
Characterization of the GPS set-up. A, Multiplexing of GPS, as illustrated for two CIMS antibodies, CIMS-17-E02 and CIMS-33–3D-F06, which were mixed and subsequently exposed to tryptic digests from human colon tissue or yeast cell lysates, respectively and then analyzed by LC-MS/MS. The statistics of the number of bound human or yeast peptides or proteins per CIMS antibody are shown. B, Peptide identification reproducibility of the complete GPS-setup, including both affinity capture and LC-MS/MS as illustrated for CIMS-33–3D-F06 exposed to human tryptic colon digest or yeast cell lysates. All peptides identified with fixed or variable modifications were excluded. C, Reproducibility of the entire GPS setup for label-free quantitative LC-MS/MS, as illustrated for CIMS-33–3D-F06 exposed to different amounts of human tryptic colon digests or yeast cell lysates. Raw abundance values (nonnormalized area under the peak values generated by Progenesis LC-MS software) are plotted. Data plotted were limited to peptides containing the experimentally refined binding motif (the frequency of the last six C-terminal amino acids is indicated for the peptides plotted). D, Dynamic range of the GPS set-up, illustrated for six CIMS antibodies immobilized on magnetic beads and exposed to tryptic digest of yeast cell lysates. The identified peptides (proteins) were plotted against the á priori known protein abundance levels of all individual yeast proteins (39). Among the detected peptides, four are indicated for two CIMS antibodies each, CIMS-17-E02 and CIMS-33–3D-F06, demonstrating the dynamic coverage. Eight detected peptides for which reference values are missing (39) (not plotted) and for which the abundance is anticipated to be <50 protein copies/cell are shown.