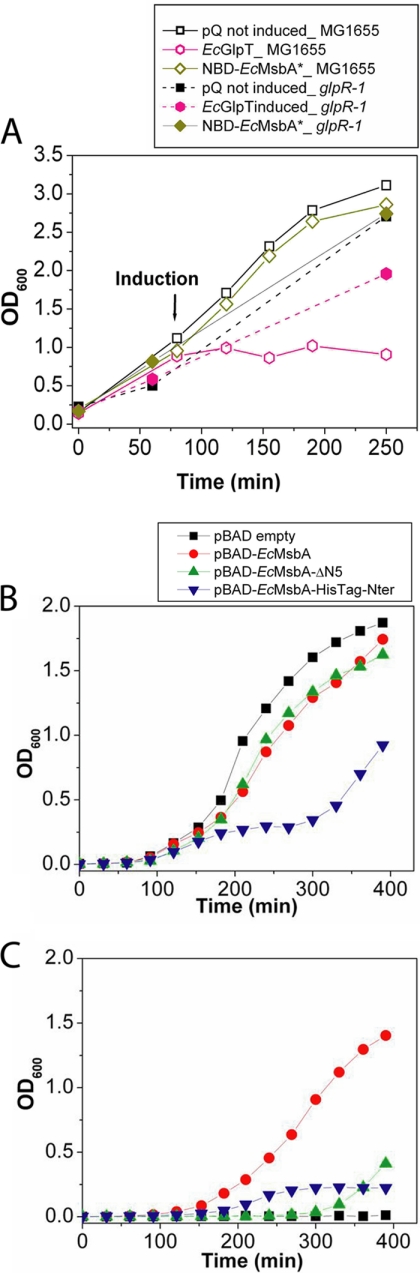
Fig. 7.
Toxicity is controlled by the biochemical and biophysical properties of the target IMP. A, OD600 was used to monitor cell growth during induction of expression of EcGlpT or NBD-EcMsbA in E. coli MG1655 cells without (empty symbols) or with (filled symbols) the glpR-1 mutation that produces constitutive expression of the enzymes mediating glycerol degradation. These experiments employed the same pQE-derived expression vectors used in Fig. 1 and elsewhere in this paper. B, Cell growth was monitored during induction of several different EcMsbA constructs from arabinose-controlled pBAD plasmids in E. coli W3110A cells: pBAD-EcMsbA expresses the full-length protein (red); pBAD-EcMsbA-ΔN5 expresses the ΔN5 construct which is missing residues 2–5 and equivalent to the EcMsbA* construct in the pQE60 vector (green); and EcMsbA-HisTag-Nter expresses the full-length protein with an N-terminal hexahistidine tag (blue). Induction was carried out at 42 °C in presence of 0.02% arabinose. C, Growth of E. coli strain WD2S containing the indicated pBAD expression plasmid at 42 °C, the nonpermissive temperature for the temperature-sensitive mutation in the chromosomally encoded msbA gene in this derivative of strain W3110A. For the experiments in panels B and C, the growth medium contained 0.02% arabinose to induce MsbA construct expression starting at zero time.