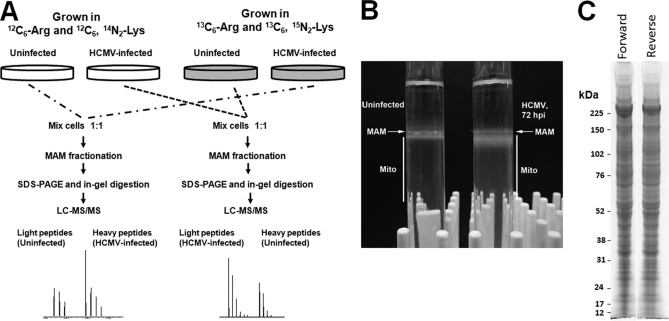
Fig. 1.
Experimental design for quantitative proteomics of enriched heavy MAM fraction from uninfected and HCMV-infected HFFs. A, Forward (left) and reverse (right) SILAC approaches. HFFs were grown in light (12C6-Arg and 12C6, 14N2-Lys) or heavy (13C6-Arg and 15N2, 13C6-Lys) medium for four cell doublings. HFFs, grown in heavy medium, were HCMV (strain BAD wt)-infected for 72 h; whereas, HFFs, grown in light medium, were uninfected for 72 h. Prior to subcellular fractionation, two roller bottles of SILAC-labeled, HCMV-infected HFFs were mixed (1:1) with two roller bottles of unlabeled, uninfected HFFs (forward experiment). In the reverse experiment, SILAC-labeled, uninfected HFFs were mixed (1:1) with unlabeled, HCMV-infected HFFs at 72 hpi. The mixed cells were fractionated to obtain heavy MAM as previously described (28, 47). Heavy MAM proteins (100 μg), obtained from mixed unlabeled and SILAC-labeled cells, were resolved by SDS-PAGE. Fifty sequential slices of each lane were in-gel digested with trypsin and analyzed by LC-MS/MS analysis. B, Percoll gradients of uninfected and HCMV infected HFFs. Uninfected HFFs and HCMV-infected HFFs (∼5 × 107 cells/each) were gently lysed by homogenization, pelleted by centrifugation at 10,300 × g for 10 min at 4 °C. The supernatant was removed and alternately processed, whereas the pellet was resuspended in 300 μl of ice-cold mannitol buffer A, briefly homogenized, layered over 10 ml of a 30% Percoll suspension in mannitol buffer B, and subjected to ultracentrifugation at 95,000 × g for 65 min at 4 °C. Banded MAM and mitochondria were recovered from the gradient by needle extraction from above (MAM, 200 μl) and below (Mito, 2 ml) the indicated points. C, SDS-PAGE of forward and reverse MAM samples. Heavy MAM from the paired forward or reverse paired HFFs were resolved by SDS-PAGE and stained by BioSafe Coomassie.