Table 3. Specificity of scoring matrices: Blast searches against a data set of membrane proteins with other architecture and a data set of globular proteins (oMBp/Globular).
| e-value | bbTMall | bbTMin | bbTMout | Blosum62 | Phat7573 | Slim161 | Pam250 |
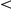 10 10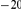
|
0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
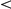 10 10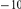
|
0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
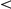 10 10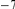
|
0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
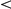 10 10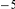
|
0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 2/2 | 0/0 |
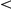 10 10
|
0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 21/3 | 0/0 |
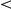 10 10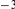
|
0/0 | 0/0 | 0/0 | 0/0 | 0/1 | 98/5 | 1/1 |
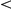 10 10
|
0/0 | 0/0 | 0/0 | 0/0 | 3/6 | 457/13 | 3/2 |
 10 10
|
0/0 | 0/0 | 0/0 | 0/0 | 13/26 | 1780/42 | 28/31 |
Cumulative number of sequences of membrane proteins with other architecture and globular protein sequences incorrectly identified as homologs of 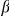 -barrel membrane proteins at different
-barrel membrane proteins at different  -values resulting from Blast searches against the oMBp/Globular data set. The number of sequences part of oMBp is 10,951 (1,061 from Archaea and 9,890 from Eukaryota). The size of the data set Globular is 127,485 globular protein sequences (16,814 Archaea and 110,671 Eukaryota). We used as queries the concatenated transmembrane sequences of the 20 template proteins.
-values resulting from Blast searches against the oMBp/Globular data set. The number of sequences part of oMBp is 10,951 (1,061 from Archaea and 9,890 from Eukaryota). The size of the data set Globular is 127,485 globular protein sequences (16,814 Archaea and 110,671 Eukaryota). We used as queries the concatenated transmembrane sequences of the 20 template proteins.
