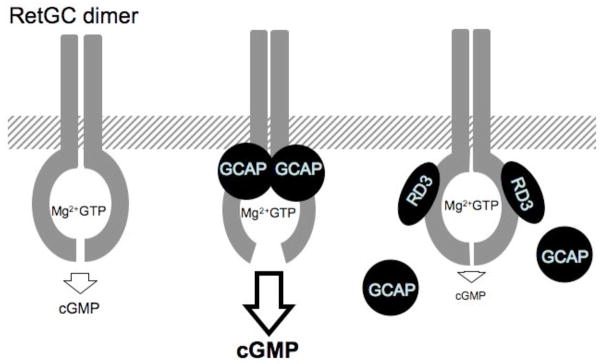
Fig. 6.
A diagram depicting the inhibition of RetGC by RD3. A RetGC homodimer forms an active site by combining the Mg2+-coordinating center from one subunit with the GTP binding center from another subunit (27–29). There are two symmetrical active sites in the dimer, but only one such site is depicted here for simplicity. The low RetGC basal catalytic activity (left) becomes accelerated up to 20–100 fold (21, 31) when GCAPs bind the cytoplasmic portion of RetGC (middle). RD3 binding to the cyclase (right) inhibits the RetGC catalytic activity, but does not prevent binding of the Mg2+GTP substrate in the active site; at the same time, RD3 acts as a negative high-affinity modulator of the RetGC/GCAP complex by displacing GCAP – this prevents dynamic activation by GCAPs via Ca2+-feedback mechanism. The GCAP-stimulated activity is much higher than the basal RetGC activity (21, 31); therefore, the strongest effect of RD3 as a negative allosteric modulator of the cyclase results from its competition with GCAP. Other explanations are provided in the Discussion.