Fig. 5.
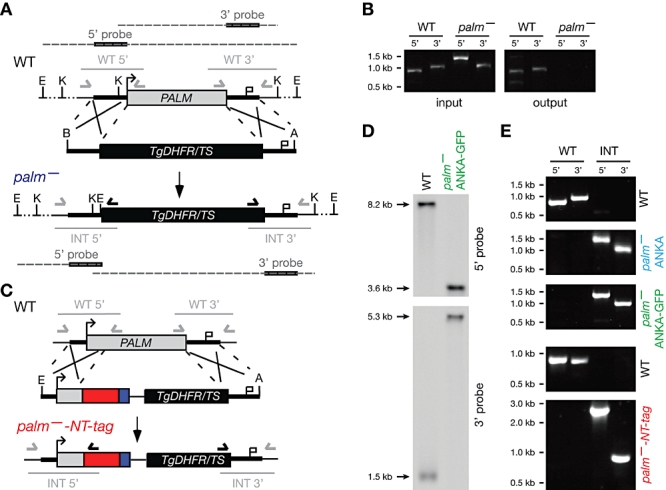
Generation of palm- parasites. A. The PbPALM genomic locus was targeted with a replacement plasmid containing PALM 5′ and 3′ fragments (thick black lines) and the DHFR/TS positive selectable marker (black box). Upon a double cross-over event, the targeting plasmid is expected to replace the endogenous PALM ORF. Arrows and light grey solid lines indicate specific primers and PCR fragments respectively. Black bars indicate specific 5′ and 3′ probes; dark grey dashed lines represent EcoRI or KpnI restriction-digested fragments of WT or knockout parasite gDNA. B. Mouse-mosquito-mouse transmission experiment starting from a mixed WT/palm- infection (input) and resulting in a pure WT population after completion of one transmission cycle (output). Parasites were genotyped by PCR using wild-type (WT)- and replacement (INT)-specific primer pairs, as indicated in (A). C. Alternative gene replacement vector to ablate the PALM locus. The PbPALM genomic locus was targeted with a replacement plasmid containing a PALM 5′ fragment, containing a portion of the PALM open reading frame (grey box), fused to the mCherry protein (red box) and quadruple myc tag (blue box), followed by the DHFR/TS positive selectable marker (black box) and the 3′ flanking region (thick bar). Upon a double cross-over event, the targeting plasmid is expected to replace the endogenous PALM ORF. Arrows and bars indicate specific primers and PCR fragments respectively. D. Genotyping of the palm- ANKA-GFP parasite line used for the majority of experiments through Southern blot analysis. Probes at the 5′ and 3′ UTR of PALM were used to demonstrate the expected size shift of restriction-digested gDNA of WT and knockout parasites. E. Genotyping of the three clonal palm- parasite lines obtained from independent transfection experiments. Using integration-specific primer combinations (A, C) the successful replacement event (INT) was verified. Absence of the WT signal from palm- parasites confirmed the purity of the clonal populations. The clonal populations palm- ANKA (blue) and palm- ANKA-GFP (green) were obtained according to design (A), using WT-ANKA and ANKA-GFP parasites as recipient lines (left); the clonal population palm--NT-tag (red) was obtained according to design (C), using ANKA-GFP parasites as recipient lines (right).
