Figure 3.
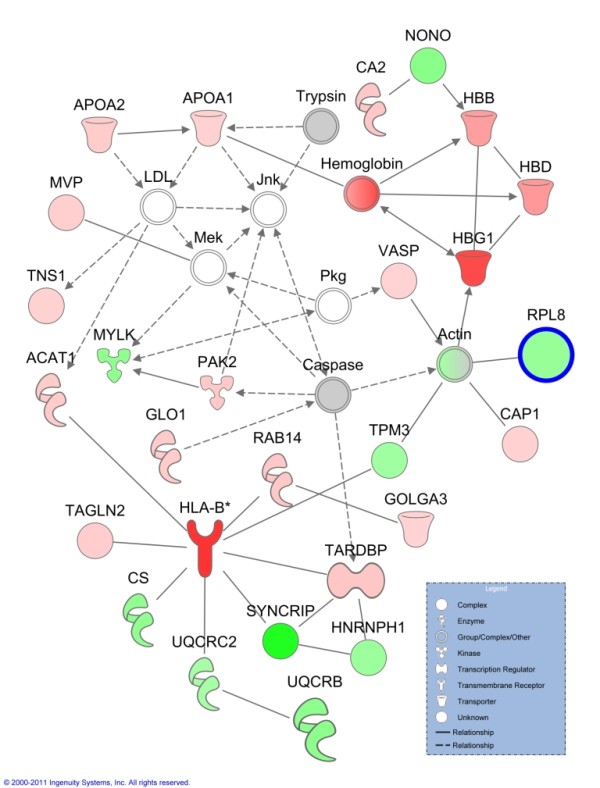
Network of differentially expressed proteins in untreated asthmatics compared to healthy controls. The top network generated by Ingenuity Pathways Analysis from quantified proteins (1.5 fold change threshold) of untreated asthmatics compared to healthy controls, contains the functions of haematological system development and function, lipid metabolism and molecular transport. Molecules with bold outline are significantly different between the two groups using a Student's t-test. Lines indicate relationships between molecules. Arrows at the end of these lines indicated the direction of the interaction. Molecules that are up-regulated and down-regulated in the dataset are coloured red and green respectively. Grey molecules do not meet the cut-off threshold. Uncoloured molecules have been added from the Ingenuity Knowledge Base. Entrez gene names for molecules are: ACAT1 = acetyl-CoA acetyltransferase 1, APOA1 = apolipoprotein A-I, APOA2 = apolipoprotein A-II, CA2 = carbonic anhydrase II, CAP1 = CAP, adenylate cyclase-associated protein 1 (yeast), CS = citrate synthase, GLO1 = glyoxalase I, GOLGA3 = golgin A3, HBB = hemoglobin (beta), HBD = hemoglobin (delta), HBG1 = hemoglobin (gamma A), HLA-B = major histocompatibility complex, class I, B, HNRNPH1 = heterogeneous nuclear ribonucleoprotein H1 (H), MVP = major vault protein, MYLK = myosin light chain kinase, NONO = non-POU domain containing, octamer-binding, PAK2 = p21 protein (Cdc42/Rac)-activated kinase 2, RAB14 = RAB14, member RAS oncogene family, RPL8 = ribosomal protein L8, SYNCRIP = synaptotagmin binding, cytoplasmic RNA interacting protein, TAGLN2 = transgelin 2, TARDBP = TAR DNA binding protein, TNS1 = tensin 1, TPM3 = tropomyosin 3, UQCRB = ubiquinol-cytochrome c reductase binding protein, UQCRC2 = ubiquinol-cytochrome c reductase core protein II, VASP = vasodilator-stimulated phosphoprotein.
