Figure 5.
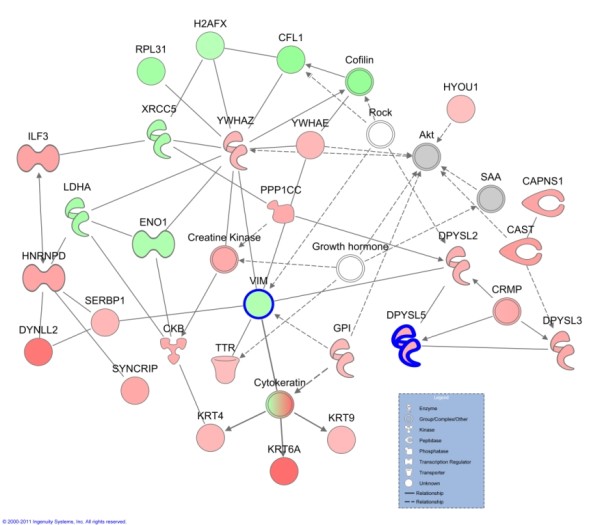
Network of differentially expressed proteins of post budesonide compared to paired pre treatment biopsy. The top network generated by Ingenuity Pathways Analysis from quantified proteins of post budesonide asthmatics compared to their pre placebo biopsy. The network contains the functions of cancer, genetic disorder and respiratory disease. Molecules with bold outline are significantly different between the two groups using a Student's t-test. Lines indicate relationships between molecules. Arrows at the end of these lines indicated the direction of the interaction. Molecules that are up-regulated and down-regulated in the dataset are coloured red and green respectively. Grey molecules do not meet the cut-off threshold. Uncoloured molecules have been added from the IPA KB. Entrez gene names for molecules are: CAPNS1 = calpain, small subunit 1, CAST = calpastatin, CFL1 = cofilin 1 (non-muscle), CKB = creatine kinase, brain, DPYSL2 = dihydropyrimidinase-like 2, DPYSL3 = dihydropyrimidinase-like 3, DPYSL5 = dihydropyrimidinase-like 5, DYNLL2 = dynein, light chain, LC8-type 2, ENO1 = enolase 1, (alpha), GPI = glucose-6-phosphate isomerase, H2AFX = H2A histone family, member X, HNRNPD = heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37 kDa), HYOU1 = hypoxia up-regulated 1, ILF3 = interleukin enhancer binding factor 3, 90 kDa, KRT4 = keratin 4, KRT9 = keratin 9, KRT6A = keratin 6A, LDHA = lactate dehydrogenase A, PPP1CC = protein phosphatase 1, catalytic subunit, gamma isozyme, RPL31 = ribosomal protein L31, SERBP1 = SERPINE1 mRNA binding protein 1, SYNCRIP = synaptotagmin binding, cytoplasmic RNA interacting protein, TTR = transthyretin, VIM = vimentin, XRCC5 = X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining), YWHAE = tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide, YWHAZ = tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide.
