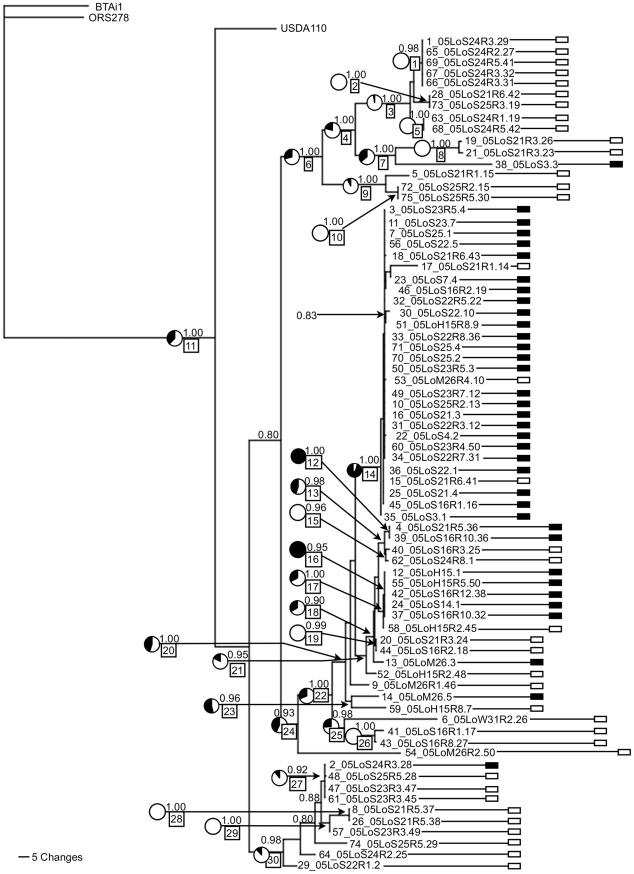
Figure 1. Ancestral state reconstruction of nodulation ability on B. japonicum phylogeny.
Bayesian phylogram of 74 Bradyrhizobium japonicum isolates from Bodega Marine Reserve [5] inferred with three loci (Its, GlnII, RecA; total 2,238 nt) and rooted with three fully sequenced Bradyrhizobium strains (B. japonicum USDA110, Bradyrhizobium sp. BTAi1 and Bradyrhizobium sp ORS278). The tree represents a single sample from the post-burnin set of trees, in which branch lengths are scaled to indicate number of nucleotide changes. Beginning from the left, taxon labels for rhizobial isolates consist of strain number (1–75), year of isolation (05 = 2005), host species (LoA = Lotus angustissimus, LoM = L. micranthus, LoH = L. heermannii, LoS = L. strigosus), plant number, and nodule or root-surface number (the latter with R followed by root and isolate number). Strain number 27 was too divergent to include on the tree as it is more closely related to Methylobacterium [22]. Symbiotic phenotypes on L. strigosus from the inoculation assays are indicated on the tips of the tree with rectangular labels (black = nodulating on L. strigosus, white = non-nodulating on L. strigosus; [22]). Bayesian clade support values (posterior probabilities) are reported above the branches of all well-supported clades (pp≥0.80). Ancestral states are estimated for all well-supported internal nodes (pp≥0.90; labeled #s 1–30 in boxes) for the binary character of nodulating or non-nodulating on L. strigosus (using parsimony, maximum likelihood and Bayesian stochastic character mapping; Table 2). Bayesian posterior probabilities of the ancestral states are reported using pie charts with black filling indicating the posterior probability of the ancestor being nodulating. In the parsimony analysis all 30 well-supported ancestral nodes were inferred to be non-nodulating except for #'s 12, 14 and 16.