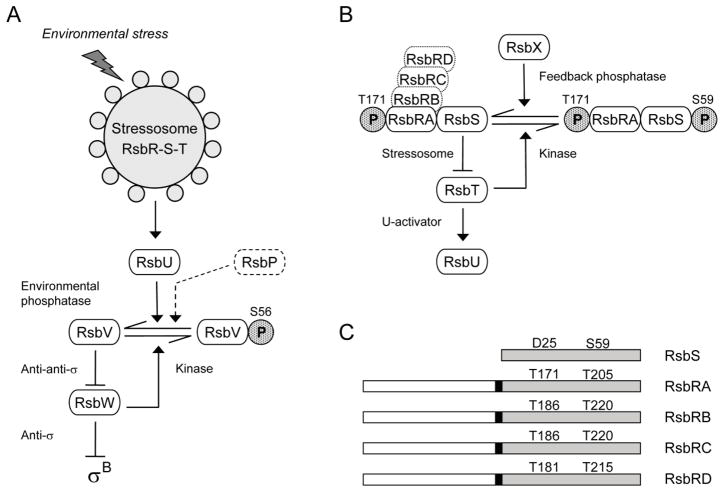
Figure 1. The σB regulatory network.
(A) Model of signaling pathways that regulate σB (modified from Hecker et al., 2007; Price, 2010). Environmental and energy pathways converge on RsbV and RsbW, which directly regulate σB activity (see text). The stressosome controls activation of the RsbU environmental phosphatase in response to diverse signals; the RsbP energy phosphatase is represented only in dotted outline. Horizontal arrows show conversion between RsbV and RsbV-P (with phosphate as stippled P). Full arrowheads indicate activating effects and T-headed lines inhibiting ones. (B) Model of stressosome control of RsbU phosphatase activity. The stressosome comprises the RsbRA, RB, RC, and RD co-antagonists and the RsbS antagonist, which together bind the RsbT kinase. In unstressed cells RsbT phosphorylates RsbRA on T171; we establish here that RsbT is also required for phosphorylation of RB on T186, RC on T186, and RD on T181. During the stress response, RsbRA-T171-P enhances the activity of the RsbT kinase, which inactivates its RsbS antagonist by phosphorylating S59; RsbT is released to bind and activate the RsbU environmental phosphatase. The RsbX feedback phosphatase dephosphorylates RsbS, damping continued signaling. (C) The RsbS antagonist consists of a single STAS domain (shaded), whereas the RsbR co-antagonists have an N-terminal, non-heme globin domain (open) connected to a C-terminal STAS domain by a 13 residue linker (black rectangle). Conserved threonine residues are indicated in the STAS domains of the RsbR proteins, together with the corresponding aspartate and serine residues of RsbS. These threonine and serine residues are the known or presumed sites of phosphorylation by RsbT.