Figure 2.
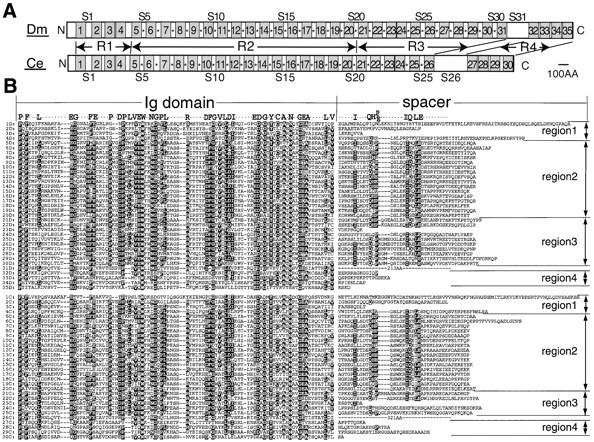
(A) Highly repetitive structures of Drosophila melanogaster (Dm) and C. elegans (Ce) Kettin proteins are schematically shown. Drosophila and C. elegans Kettins contain numerous repeating units consisting of an Ig domain and a spacer sequence. In both Drosophila and C. elegans, Kettin can be divided into four regions, R1-4, except for the NH2-terminal region and a long spacer (S31 and S26, respectively, in Drosophila and C. elegans). Stippled boxes with numbers show Ig domains. S1–S31 indicate spacer numbers. Spacers with conserved Kettin spacer motifs are marked with asterisks. (B) Comparison of Ig domain amino acid sequences of Drosophila and worm Kettins. iD (i = 1–35) indicates the Ig domain and spacer sequences of the ith repeating unit of Drosophila Kettin, while iC (i = 1–30), those of the ith repeating unit of worm Kettin. Ig domains consist of ∼95 amino acids. Highly conserved amino acids are labeled by white letters in black. Note that spacers in regions 2 and 3 share in common two conserved Kettin spacer motifs, I (IXXXQH(P/E)) and II (IQXLE).
