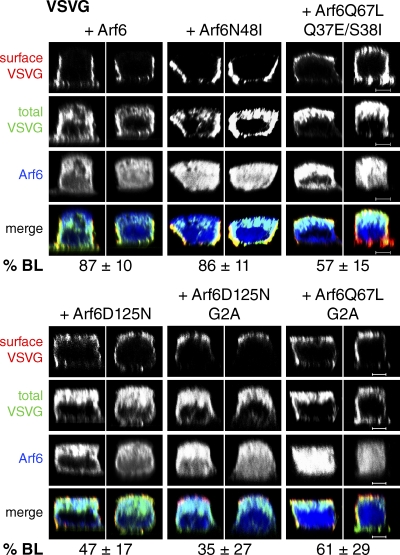
Figure 4.
Analysis of VSVG sorting in the presence of various Arf6 mutant proteins. Polarized, filter-grown MDCK cells were coinjected with plasmids encoding VSVGts045-GFP and V5-tagged Arf6 proteins: wild-type Arf6, Arf6N48I, Arf6Q67L/Q37E/S38I, Arf6D125N, Arf6D125N/G2A, or Arf6Q67L/G2A. Cells were processed as described for VSVG (Fig. 2). Specimens were analyzed by confocal microscopy, and representative xz sections are shown. Numerical data were obtained by using Volocity software, as described in Materials and methods, from at least three independent experiments. For all conditions, numerical data for the mock control were the same as in Fig. 2 A. P-values are in comparison to the mock control unless stated otherwise. Errors indicate SD. Arf6: n = 29, P = 0.0234. Arf6N48I: n = 39, P = 0.0079. Arf6Q67L/Q37E/S38I: n = 31, P < 0.0001 in comparison to both Arf6Q67L and mock control. Arf6D125N: n = 35, P < 0.0001. Arf6D125N/G2A: n = 35, P < 0.0001 and 0.0294 in comparison to mock control and Arf6D125N, respectively. Arf6Q67L/G2A: n = 29, P < 0.0001 and 0.0014 in comparison to mock control and Arf6Q67L, respectively. Bars, 5 µm.