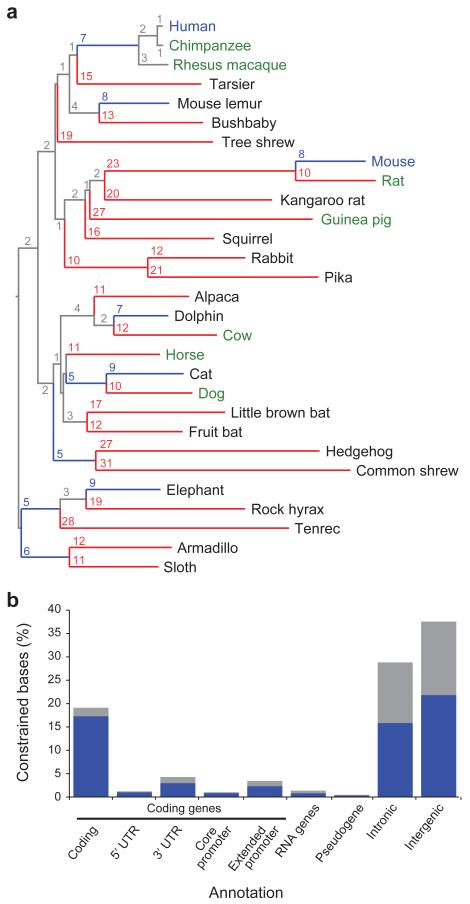
Figure 1. Phylogeny and constrained elements from the 29 eutherian mammalian genome sequences.
a, A phylogenetic tree of all 29 mammals used in this analysis based on the substitution rates in the MultiZ alignments. Organisms with finished genome sequences are indicated in blue, high quality drafts in green and 2X assemblies in black. Substitutions per 100 bp are given for each branch, and branches with ≥ 10 substitutions are colored red, while blue indicates < 10 substitutions. b, At 10% FDR, 3.6 million constrained elements can be detected encompassing 4.2% of the genome, including a substantial fraction of newly detected bases (blue) compared to the union of the HMRD 50-bp + Siepel vertebrate elements17 (see Figure S4b for comparison to HMRD elements only). The largest fraction of constraint can be seen in coding exons, introns and intergenic regions. For unique counts, the analysis was performed hierarchically: coding exons, 5′-UTRs, 3′-UTRs, promoters, pseudogenes, non-coding RNAs, introns, intergenic. The constrained bases are particularly enriched in coding transcripts and their promoters (Supp Fig S4c).