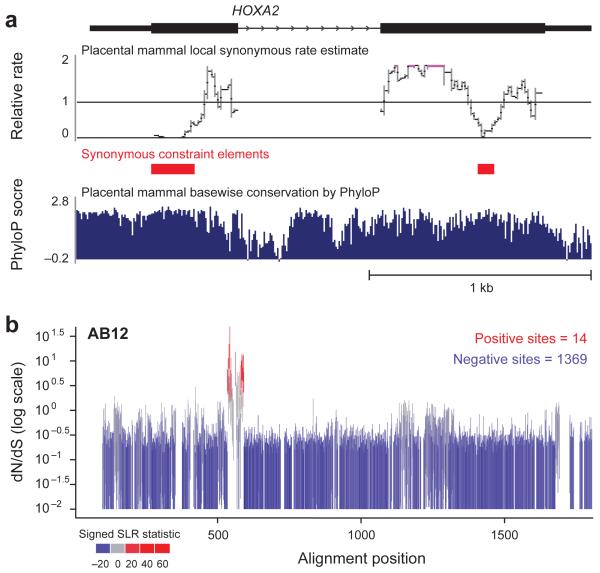
Figure 3. Examination of evolutionary signatures identifies synonymous constrained elements (SCEs) and evidence of positive selection.
a, Two regions within the HOXA2 open reading frame are identified as Synonymous Constraint Elements (red), corresponding to overlapping functional elements within coding regions. Note that the synonymous rate reductions are not obvious from the base-wise conservation measure (in blue). Both elements have been characterized as enhancers driving Hoxa2 expression in distinct segments of the developing mouse hindbrain. The element in the first exon encodes Hox-Pbx binding sites and drives expression in rhombomere 433, while the element in the second exon contains Sox binding sites and drives expression in rhombomere 232. Synonymous constraint elements are also found in most other Hox genes, and up to a quarter of all genes. b, While ~85% of genes show only negative (purifying) selection and 9 % of genes show uniform positive selection, the remaining 6% of genes, including ABI2, show only localized regions of positively-selected sites. Each vertical bar covers the estimated 95% confidence interval for dN/dS at that site (with values of 0 truncated to 0.01 to accommodate the log scaling), and bars are colored according to a signed version of the SLR statistic for non-neutral evolution: blue for sites under purifying selection, gray for neutral sites, and red for sites under positive selection.