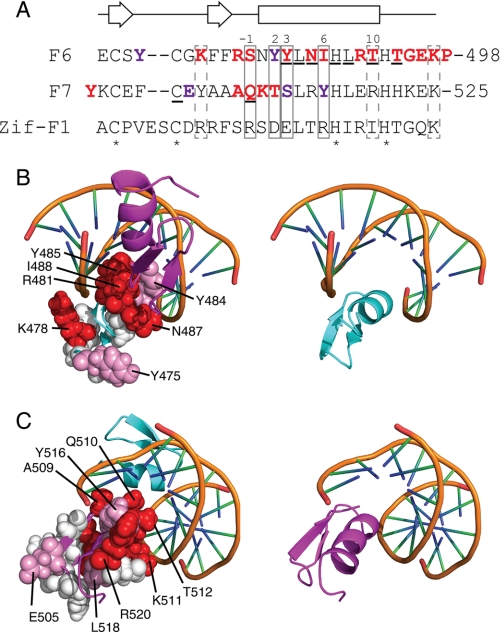
FIGURE 10.
Residues important for ZNF217-F67 DNA recognition. A, the amino acid sequences of F6 and F7 of ZNF217 are shown together with the sequence of finger 1 from the prototypical DNA binding zinc finger protein Zif268. Residues that typically make sequence-specific contacts with DNA in classical zinc fingers are boxed with solid lines, residues that often make nonspecific interactions with the DNA backbone are shown in dashed boxes, residues that underwent substantial chemical shift changes upon the addition of DNA to ZNF217-F67 are underlined, and zinc-ligating residues are indicated with asterisks. Those residues shown by site-directed mutagenesis to mostly or completely eliminate DNA binding are in bold and colored red; residues that reduced but did not abolish DNA binding are in bold and colored purple. Numbering of the α-helix is that typically used for classical zinc fingers. The secondary structure for F6 and F7, as predicted from an analysis of F67 chemical shifts, is shown above the sequences. B and C, the structures of F6 and F7 were overlaid onto F2 and F3 of Zif268 in the x-ray crystal structure of this protein bound to DNA (PDB 1ZAA). Residues that were mutated in Fig. 8 are shown in space-filling representation. Those residues shown by site-directed mutagenesis to mostly or completely eliminate DNA binding are colored red, residues that reduced but did not abolish DNA binding are colored pink, and residues that had little or no effect on DNA binding are shown in white. Red residues are more prominent on the surface of the domains used in Zif268 to contact DNA, whereas white residues are concentrated on the opposite face of each domain. B shows residues in F6, whereas C shows residues in F7. The right-hand panels show F6 or F7 in the same orientation as the corresponding left-hand panel for reference.