TABLE 2.
Summary of GlgE x-ray data for refinement and model parameters
r.m.s.d. is root mean square deviation and NA is not applicable.
| Data set | Apo-GlgE | αCD-GlgE | mal-GlgE | αCD-mal-GlgE | βCD-mal-GlgE |
|---|---|---|---|---|---|
| Data collection | |||||
| Space groupa | P212121 | P41212 | P212121 | P212121 | P212121 |
| Cell parameters | a = 113.1, b = 113.0, c = 314.2 Å | a = b = 113.2, c = 314.5 Å | a = 113.8, b = 113.6, c = 315.7 Å | a = 113.9, b = 114.2, c = 315.6 Å | a = 113.3, b = 113.4, c = 315.0 Å |
| Beamlineb | I03 | I02 | I04 | I04 | I02 |
| Wavelength | 0.9709 Å | 0.9795 Å | 0.9763 Å | 0.9763 Å | 0.9795 Å |
| Resolution rangec | 53.15 to 1.80 Å | 71.35 to 2.30 Å | 56.89 to 2.10 Å | 63.99 to 2.20 Å | 71.42 to 2.50 Å |
| (1.90 to 1.80 Å) | (2.42 to 2.30 Å) | (2.21 to 2.10 Å) | (2.32 to 2.20 Å) | (2.64 to 2.50 Å) | |
| Unique reflectionsc | 351,739 | 85,772 | 236,917 | 206,349 | 140,547 |
| Completenessc | 95.0% (72.9%) | 93.7% (66.0%) | 99.5% (96.7%) | 98.9% (93.2%) | 99.8% (99.7%) |
| Redundancyc | 7.6 (5.5) | 14.3 (13.2) | 7.3 (4.7) | 5.4 (4.1) | 4.9 (5.0) |
| Rmergec,d | 0.110 (0.762) | 0.088 (0.327) | 0.121 (0.426) | 0.151 (0.602) | 0.143 (0.688) |
| Rmeasc,e | 0.118 (0.840) | 0.092 (0.340) | 0.130 (0.482) | 0.167 (0.692) | 0.160 (0.767) |
| Mean I/σ(I)c | 13.4 (2.1) | 23.6 (7.9) | 12.0 (3.2) | 7.7 (2.1) | 10.0 (2.3) |
| Wilson B value | 21.6 Å2 | 33.2 Å2 | 26.1 Å2 | 33.8 Å2 | 42.4 Å2 |
| Twin fractionf | 0.39 | 0.04 | 0.36 | 0.27 | 0.17 |
| Refinement | |||||
| Reflections: working/freeg | 333,972/17,659 | 81,357/4,301 | 224,955/11,847 | 195,790/10,447 | 133,513/6,958 |
| Rworkh | 0.231 | 0.173 | 0.204 | 0.208 | 0.191 |
| Rfreeh | 0.249 | 0.201 | 0.228 | 0.236 | 0.221 |
| Ramachandran favored/allowedi | 98.6/100.0% | 98.7/100.0% | 98.8/100.0% | 98.7/100.0% | 98.5/100.0% |
| Ramachandran outliersi | 0 | 0 | 0 | 0 | 0 |
| r.m.s.d. bond distances | 0.013 Å | 0.016 Å | 0.015 Å | 0.015 Å | 0.014 Å |
| r.m.s.d. bond angles | 1.34° | 1.53° | 1.44° | 1.46° | 1.45° |
| Twin fractionj | 0.48 | NA | 0.49 | 0.45 | 0.49 |
| Contents of model | |||||
| Protein residues | 4 × 649 | 2 × 649 | 4 × 649 | 4 × 649 | 4 × 649 |
| Glucans | 0 | 2 × αCD | 4 × mal | 4 × αCD; 4 × mal | 4 × βCD; 4 × mal |
| Ethylene glycol | 0 | 1 | 0 | 0 | 0 |
| Water molecules | 540 | 731 | 486 | 459 | 369 |
| Average atomic displacement parameters (Å2) | |||||
| Main chain atoms | 25.0 | 47.1 | 28.6 | 32.7 | 31.8 |
| Side chain atoms | 25.6 | 49.2 | 29.1 | 33.9 | 32.5 |
| Glucans | 78.5 | 25.1 | αCD: 69.3; mal: 28.3 | βCD: 41.9; mal: 37.5 | |
| Ethylene glycol | 49.8 | ||||
| Water molecules | 20.2 | 44.9 | 21.7 | 24.4 | 21.4 |
| Overall | 25.2 | 48.3 | 28.7 | 33.5 | 32.1 |
| PDB accession code | 3zss | 3zst | 3zt5 | 3zt6 | 3zt7 |
a Space group that was used for refinement.
b I02, I03, I04 = beamlines at the Diamond Light Source (Oxfordshire, UK).
c The figures in parentheses indicate the values for outer resolution shell.
d Rmerge = 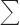 hkl
hkl 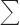 i|Ii(hkl) − 〈I(hkl)〉/
i|Ii(hkl) − 〈I(hkl)〉/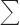 hkl
hkl 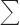 iIi(hkl), where Ii(hkl) is the ith observation of reflection hkl, and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
iIi(hkl), where Ii(hkl) is the ith observation of reflection hkl, and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
e Rmeas = 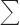 hkl [N/(N − 1)]1/2
hkl [N/(N − 1)]1/2 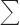 i|Ii(hkl) − 〈I(hkl)〉|/
i|Ii(hkl) − 〈I(hkl)〉|/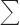 hkl
hkl 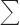 iIi(hkl), where N is the number of observations of reflections hkl.
iIi(hkl), where N is the number of observations of reflections hkl.
f Data were as calculated by TRUNCATE (24).
g The data sets were split into “working” and “free” sets consisting of 95 and 5% of the data, respectively. The free set was not used for refinement.
h The R-factors Rwork and Rfree are calculated as follows: R = 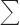 (|Fobs − Fcalc|)/
(|Fobs − Fcalc|)/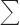 |Fobs| ×100, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
|Fobs| ×100, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
i Data were calculated using MOLPROBITY (32).
j Refined values were from REFMAC5 (29).
