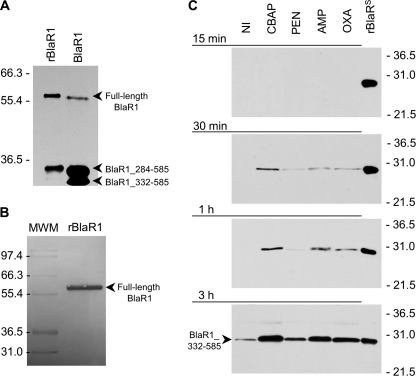
FIGURE 4.
Fragmentation of BlaR1 in CBAP-induced NRS128 and in E. coli and the shedding of its sensor domain in antibiotic-induced S. aureus NRS128. A, full-length BlaR1 in CBAP-induced NRS128 (right lane) and in E. coli (left lane) have comparable sizes, with that in E. coli larger by the size of the His6 tag moiety. BlaR1 immunoprecipitated from CBAP-induced NRS128 and from E. coli was separated by SDS-PAGE (10% gel), transferred to PVDF membrane, and detected by Western blot using the anti-BlaRS antibodies and the ONE-HOUR IP-WesternTM kit (GenScript). B, recombinant BlaR1 (∼60-kDa) obtained from E. coli whole-cell extracts by means of the preparative purification for Edman degradation, visualized in the Coomassie-stained PVDF membrane. Numbers on the left of A and B indicate the position of migration of the molecular mass markers (kDa). C, detection of the C-terminal domain of BlaR1 (BlaRS or BlaR1_332–585) in media of the S. aureus NRS128 culture after induction with different β-lactam antibiotics (CBAP, PEN, AMP, and OXA) for different times (15 min, 30 min, 1 h, and 3 h). The results for the three other strains are given in the supplemental material. The proteins were separated by SDS-PAGE (12% gels) and transferred to nitrocellulose membrane, and BlaR1 was detected by Western blot using antibodies specific to recombinant BlaRS and the ONE-HOUR IP-WesternTM kit. Numbers on the right indicate the position of migration of the molecular mass markers (kDa).