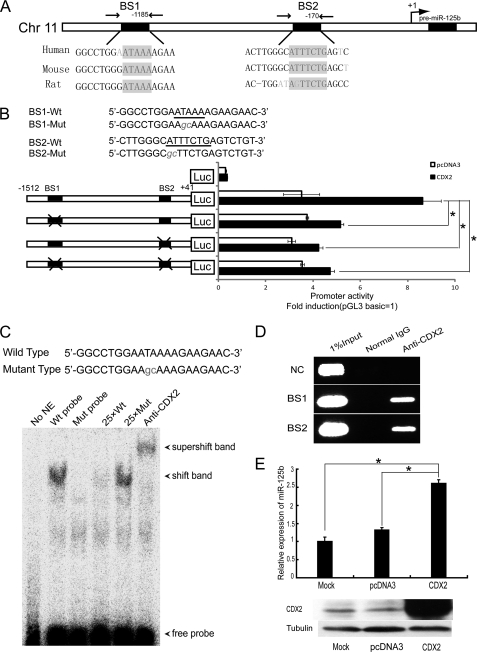
FIGURE 2.
CDX2 is responsible for miR-125b activation. A, schematic representation of the miR-125b genomic region. The sequence and location of the Cdx2-binding sites are indicated by black boxes (BS1 and BS2), whereas the arrows point to the regions amplified by PCR in the ChIP experiments. B, upper panel, mutant sequences of the miR-125b promoter reporter vector are shown. Lower panel, 293T cells were co-transfected with a CDX2 expression vector or empty vector and various luciferase reporter vectors containing the miR-125b promoter for 24 h, followed by measurement of luciferase activity. C, electrophoretic mobility shift assay showing direct binding of CDX2 to the miR-125b promoter on BS1. Nuclear extracts with wild-type (lanes 2 and 4–6) or mutant probes (lane 3) were analyzed for their ability to bind to the promoter. D, interaction of CDX2 with the putative CDX2 binding sites on the promoter region of miR-125b were examined in NB4 cells using a ChIP assay. One percent of input DNA was used as a positive control for PCR. The IgG-immunoprecipitated chromatin was used as a control for CDX2. A irrelevant sequence 5 kbp upstream of miR-125b promoter was used as a negative control (NC). E, upper panel, the level of miR-125b in NB4 cells transfected with water, pcDNA3, or CDX2 expression vector was examined by qPCR. Lower panel, the level of CDX2 in NB4 cells transfected with water, pcDNA3, or CDX2 expression vector was examined by Western blot. *, p < 0.05.