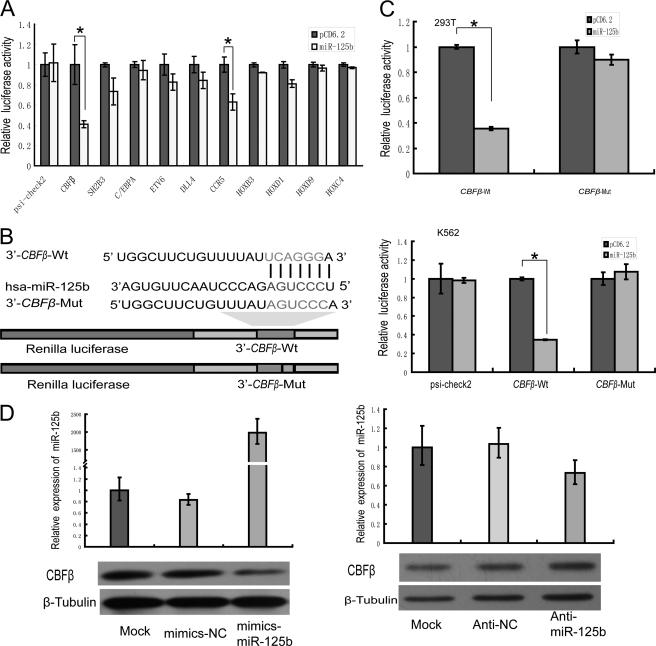
FIGURE 3.
CBFβ is a direct target of miR-125b. A, 293T cells were co-transfected putative targets of miR-125b, including the CBFβ, SH2B3, C/EBPα, ETV6, DLL4, CCR5, and HOX genes, with miR-125b expression vector (miR-125b) or control vector (pCD6.2) using Lipofectamine 2000 reagent. Renilla luciferase activity was normalized to firefly luciferase activity, and the results were expressed relative to the pCD6.2 group. The 3′-UTRs of these genes were predicted to bind to miR-125b. Only CBFβ and CCR5 showed a reduction in activity, and CBFβ showed the greatest reduction in activity (50 plus 5%). B, schematic representation of the constructs used in the luciferase assay. The sequences shown below indicate the putative miR-125b target site on the wild-type 3′-UTR (construct CBFβ-wt), its mutated derivative (construct CBFβ-mut), and the pairing regions of miR-125b. C, 293T cells (upper panel) or K562 cells (lower panel) were co-transfected psi-check2 with either CBFβ-wt or CBFβ-mut, plus either pre-miR-125b (miR-125b) or empty vector (pCD6.2). Renilla luciferase activity was expressed relative to the pCD6.2 group. D, NB4 cells were electroporated with water or 100 pmol of mimics NC (mimics-scramble), mimics-miR-125b (left panel), anti-NC (anti-scramble), or anti-miR-125b (right panel). The levels of miR-125b were examined by qPCR. Cells lysates were prepared for Western blotting with an antibody against CBFβ, and the expression of β-tubulin served as a loading control. *, p < 0.05.