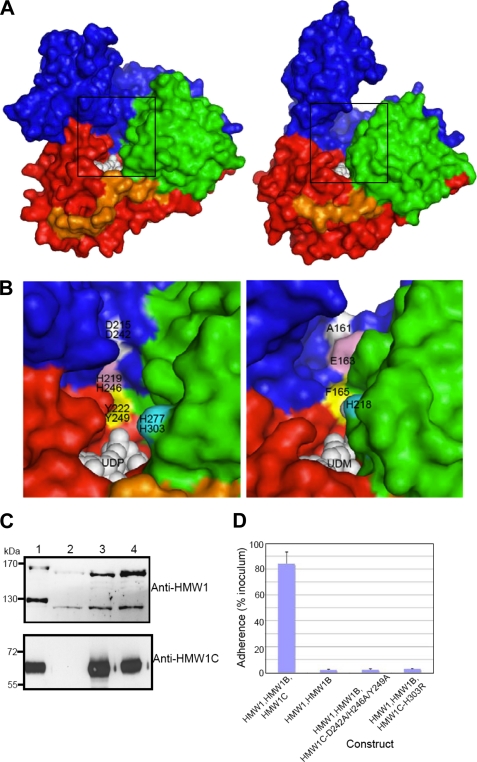
FIGURE 4.
Putative sugar/peptide-binding sites and functional implications. A, surface representation of ApHMW1C-UDP complex (left) and XcOGT-UDPGlcNAc analog (UDM) complex (right). For comparison of the putative peptide binding clefts, the ApHMW1C and XcOGT (PDB 2JLB) structures were first superimposed. Thus, the two views are from the same orientation. B, close-up view of the groove area, as indicated with boxes in A: ApHMW1C (left) and XcOGT (right). ApHMW1C and HMW1C residues were tested for activity by mutagenesis and functional assays. For clarity, representative residues are colored and labeled: ApHMW1 residues (top) and HMW1C (bottom). The corresponding groove region of XcOGT (right panel) differs from the groove in HMW1C-like proteins. C, Western immunoblots of whole cell sonicates of E. coli DH5α expressing HMW1, HMW1B, and wild type HMW1C (lane 1), HMW1 and HMW1B (lane 2), HMW1, HMW1B, and HMW1C-D242A/H246A/Y249A (lane 3), and HMW1, HMW1B, and HMW1C-H303R (lane 4). The upper blot was performed with guinea pig antiserum GP85 against HMW1, and the lower blot was performed with guinea pig antiserum GP64 against HMW1C. D, in vitro adherence assay showing HMW1-mediated adherence by E. coli DH5α expressing HMW1 and HMW1B along with wild type H. influenzae HMW1C, no HMW1C, or an HMW1C mutant.