TABLE 1.
Statistics from crystallographic analysis
| Data Collection | EMTS (Hg) | apo ApHMW1C | UDP-Glc::ApHMW1C | Peptide::ApHMW1C |
| Wavelength (Å) | 1.008 | 0.9794 | 0.9794 | 0.9794 |
| Space group | P212121 | |||
| Cell parameters (Å) | a = 79.739 | a = 80.368 | a = 80.245 | a = 79.701 |
| b = 93.888 | b = 94.677 | b = 94.896 | b = 93.262 | |
| c = 177.482 | c = 176.911 | c = 176.791 | c = 176.705 | |
| Resolution (Å) | 50.00–3.00 (3.05–3.00) | 50.00–2.10 (2.18–2.10) | 50.00–2.25 (2.33–2.25) | 50.00–2.45 (2.54–2.45) |
| Reflections (Total / Unique) | 260058 / 27518 | 262452 / 73875 | 209323 / 61133 | 183950 / 46099 |
| Completeness (%)a | 96.4 (91.5) | 93.3 (83.3) | 92.8 (74.2) | 94.1 (83.1) |
| Rmerge (%)a, c | 12.8 (61.2) | 8.0 (29.1) | 8.4 (27.1) | 8.0 (30.3) |
| Mean <I/σ(I)> | 28.6 | 12.5 | 16.5 | 13.4 |
| Redundancya | 9.5 (9.6) | 3.7 (2.4) | 3.6 (1.9) | 4.1 (2.5) |
| Sigma cutoff | 0.5 | 0.5 | 0.3 | |
| Phasing | ||||
| Overall FOMb (before / after DM) | 0.167 / 0.836 | |||
| Overall Phasing Power | 1.09 | |||
| Structure Refinement | ||||
| Resolution (Å) | 2.10 | 2.25 | 2.45 | |
| No. reflections (working/test) | 70123 / 3720 | 57402 / 3065 | 43748 / 2329 | |
| R (%)a, d/Rfreee (%) | 18.4 / 22.6 | 18.6/24.4 | 17.8 / 24.3 | |
| Number of atoms (protein/water/GOL/UDP) | 9742 / 484 / 18 / - | 9736 / 280 / 12 / 50 | 9721 / 163 / 24 / - | |
| Average B factor (protein/water/GOL/UDP, Å2) | 25.2 / 28.2 / 43.1 / - | 35.7 / 34.7 / 52.4 / 52.1 | 37.6 / 33.0 / 47.2 / - | |
| rms deviation | ||||
| Bonds (Å) | 0.016 | 0.019 | 0.021 | |
| Angles (°) | 1.607 | 1.812 | 1.933 | |
| Ramachandran plot (%) | ||||
| Most favored regions | 92.1 | 91.6 | 91.7 | |
| Additional allowed regions | 7.8 | 7.8 | 7.8 | |
| Generously allowed regions | 0.0 | 0.4 | 0.4 | |
| Disallowed regions | 0.2 | 0.2 | 0.2 | |
a Completeness and Rmerge are given for overall data and for the highest resolution shell.
b The figure of merit (FOM) = |Fbest| − |F|.
c Rmerge = 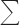 |Ii − I〉/|/sigma]|Ii|, where Ii is the intensity of an observation, I〉 is the mean value for that reflection, and the summations are overall equivalents.
|Ii − I〉/|/sigma]|Ii|, where Ii is the intensity of an observation, I〉 is the mean value for that reflection, and the summations are overall equivalents.
d R-factor = 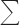 h||Fo(h)| − Fc(h)||/
h||Fo(h)| − Fc(h)||/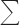 hFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
hFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
e Rfree was calculated with 5% of the data excluded from the refinement.
