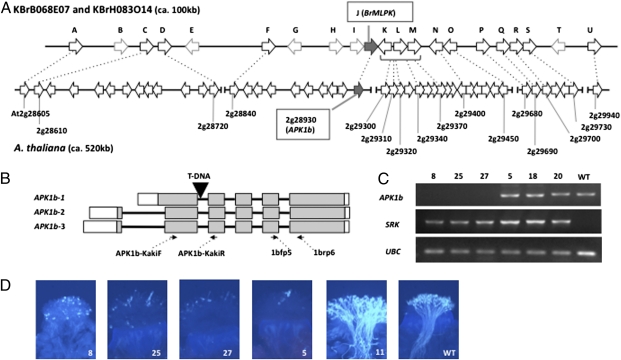
Fig. 1.
Functional analysis of a T-DNA insertion allele of AtAPK1b. (A) Synteny of the Brassica and A. thaliana chromosomal regions containing MLPK and APK1b, respectively. E-values are listed in Table S5. (B) Structure of the AtAPK1b gene showing the two alternative transcript forms produced by the gene and the location of the T-DNA insertion in the SALK_055314 strain. The boxes indicate exons; shaded boxes represent protein-coding regions, and white boxes designate untranslated regions. The arrows below the diagram indicate the location of the primers (F and R) used for RT-PCR. (C) RT-PCR analysis of stigma RNA isolated from F2 plants derived from a cross of the SALK_055314 T-DNA insertion strain and a Col-0[SRKb-SCRb] plant. Plants 8, 25, and 27 are homozygous for the T-DNA allele and do not express AtAPK1b transcripts, whereas plants 5, 18, and 20 lack the T-DNA insertion and express AtAPK1b transcripts at levels equivalent to those in WT Col-0. All of these F2 plants express SRKb transcripts. UBC transcripts were used as a control. (D) SI phenotype of F2 plants. Stigmas from stage 13 floral buds were pollinated with SCRb-expressing pollen. Plants 8, 25, and 27, which are homozygous for the T-DNA insertion in AtAPK1b and contain SRKb-SCRb, exhibit as robust an SI response as that observed in plant 5, which produces AtAPK1b transcripts and contains SRKb-SCRb. In contrast, plant 11, which is homozygous for the T-DNA allele but lacks SRKb-SCRb, did not exhibit SI, similar to WT Col-0.