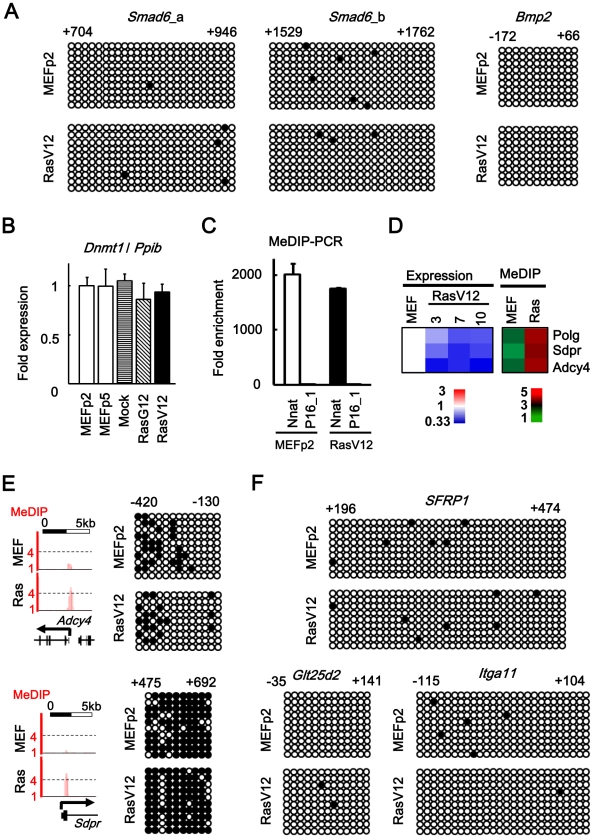
Figure 6. No detection of DNA methylation alteration.
(A) Bisulfite sequencing of 5′ regions of Smad6 and Bmp2. Positions were shown with TSS regarded as +1. Open circle, unmethylated CpG site. Closed circle, methylated CpG site. Nine to 10 clones were analyzed in each region, and aligned vertically; 10 – 36 CpG sites within the analyzed regions were aligned horizontally. Since H3K27me3 mark in Smad6 was stretched towards exon 1 and intron 1 (Figure 3A), two regions (a and b) were analyzed for Smad6. There was no DNA methylation alteration in Smad6 and Bmp2. (B) Real-time PCR for Dnmt1. Dnmt1 expression level was not altered in Ras-induced senescence, or during passages. (C) Validation of enrichment of methylated DNA in MeDIP. MeDIP-PCR was performed for 5′ regions of Nnat and p16 (region 1 in Figure 1A), and fold enrichment relative to p16 was shown. Nnat is an imprinted gene and a positive control for DNA methylation(+) region, and enrichment of methylated DNA in MeDIP was validated. (D) Analysis of MeDIP-seq. When analyzing ±1 kb of TSS of 20,232 genes, only 3 genes showed increase of MeDIP status from <2 reads per million reads in MEFp2 to >4 reads per million reads in RasV12 cells. (E) Validation of MeDIP-seq result. Among the three candidate methylated genes, Adcy4 and Sdpr were chosen for validation by bisulfite sequencing. The methylation statuses of these genes, however, were not altered in Ras-induced senescence. (F) Bisulfite sequencing for 5′ regions of other genes. Among genes showing slight increase of MeDIP status, bisulfite sequencing was performed for five chosen genes: Sfrp1 (from 0.7 reads in MEFp2 to 1.6 reads in RasV12 cells), Glt25d2 (from 0.9 to 2.5), Itga11 (0.6 to 2.7), Shisa2 (0.6 to 2.0), and Gypc (0.8 to 1.9). Sfrp1, Glt25d2, and Itga11 were representatively shown. These five genes were unmethylated in both MEFp2 and RasV12 cells.