Figure 1.
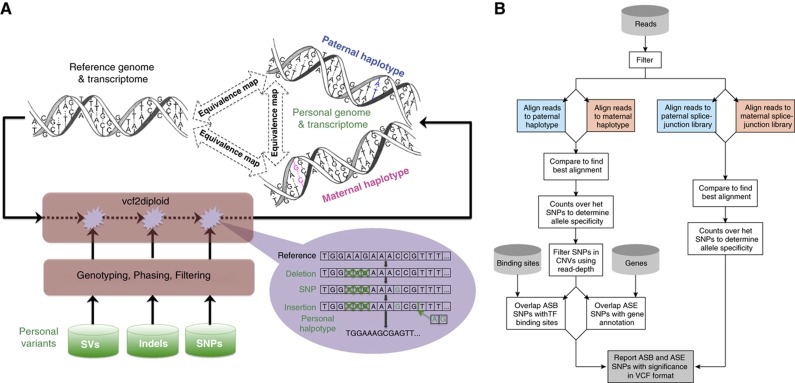
(A) Construction of a personal genome by vcf2diploid tool is made by incorporating personal variants into the reference genome. Personal variants may require additional pre-processing, that is, filtering, genotyping, and/or phasing. The output is the two (paternal and maternal) haplotypes of personal genome. During the construction step, the reference genome is represented as an array of nucleotides with each cell representing a single base. Iteratively, the nucleotides in the array are being modified to reflect personal variations. Once all the variations have been applied, a personal haplotype is constructed by reading through the array. Simultaneously, equivalence map (MAP-file format—see Supplementary Figure 1) between personal haplotypes and reference genome is being constructed. This can similarly be done for a personal transcriptome. (B) AlleleSeq pipeline for determining allele-specific binding (ASB) and allele-specific expression (ASE) aligning reads against the personal diploid genome sequence as well as a diploid-aware gene annotation file (including splice-junction library).