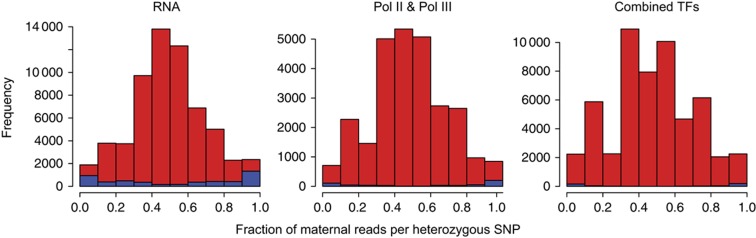
Figure 2.
For each heterozygous SNP location covered at a depth greater than six, we can compute the fraction of reads derived from the alternative allele relative to the reference sequence. We then plotted the distribution of alternative allele fraction for all heterozygous SNPs (significant allele-specific positions are indicated in blue) for the RNA-Seq, Pol II, and remaining ChIP-Seq data sets combined. We observe that the distribution of all heterozygous SNPs as well as the allele-specific SNP positions is quite symmetric; and thus, we do not see a significant reference bias.