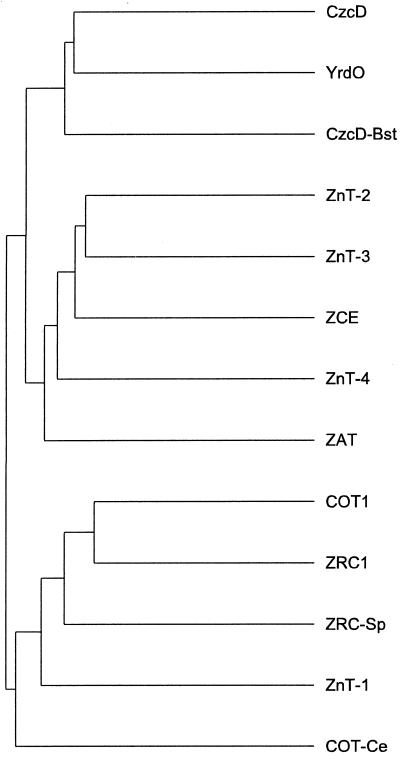
Figure 2.
Dendrogram of the amino acid sequences of several (putative) Zn transporters. CzcD, Alcaligenes eutrophus (accession no. D67044); YrdO (CzcD), Bacillus subtilis (accession no. U93876 [U62055]); CzcD-Bst, Bacillus stearothermophilus (accession no. D87026); ZnT-2, Rattus norvegicus (accession no. S70632; Palmiter et al., 1996a); ZnT-3, Homo sapiens (accession no. U76010; Palmiter et al., 1996b); ZCE, C. elegans (accession no. T18D3.3 in Z68119); ZnT-4, H. sapiens (accession no. AF025409; Huang and Gitschier, 1997); ZAT, Arabidopsis (this study); COT1, Saccharomyces cerevisiae (accession no. P32798; Conklin et al., 1992); ZRC1, S. cerevisiae (accession no. P20107; Kamizono et al., 1989); ZRC-Sp, ZRC1 homolog, Schizosaccharomyces pombe (accession no. D89236); ZnT-1, R. norvegicus (accession no. S54303; Palmiter et al., 1995); COT-Ce, COT1 homolog in C. elegans (accession no. U23529). The distance between the branches on the horizontal axis are proportional to the degree of relatedness according to the program used, whereas the distances along the vertical axis are without any meaning.