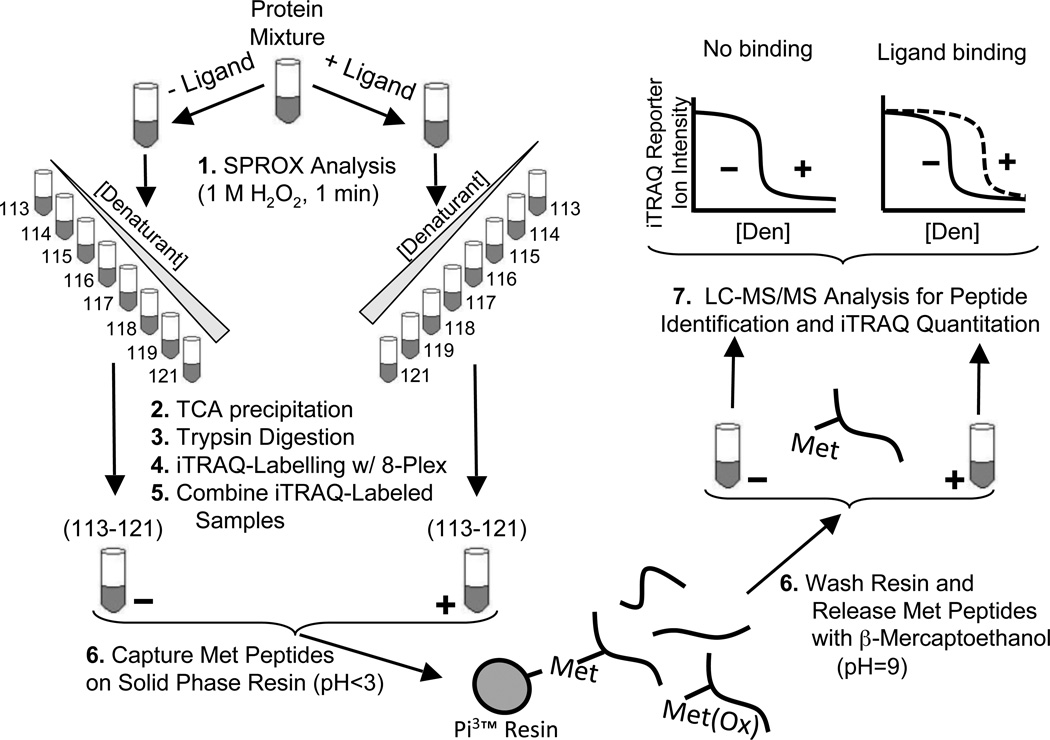
Figure 1.
Schematic representation of the protocol used in this work. A protein mixture is incubated in the presence and absence of ligand before diluting into a series of denaturant-containing buffers. Following SPROX analysis, trypsin digestion, and iTRAQ labeling with the eight different iTRAQ reagents bearing the 113 – 121 reporter ions, the samples across the range of denaturants are combined (i.e., the −ligand samples labeled with the eight iTRAQ reagents bearing the 113–121 reporter ions are combined and the +ligand samples labeled with the eight iTRAQ reagents bearing the 113–121 reporter ions are combined). Ultimately, the − and + ligand samples are each subjected to a Met peptide selection strategy. The samples enriched for Met peptides are analyzed by LC-MS/MS, and iTRAQ reporter ions are used to generate chemical denaturation data.