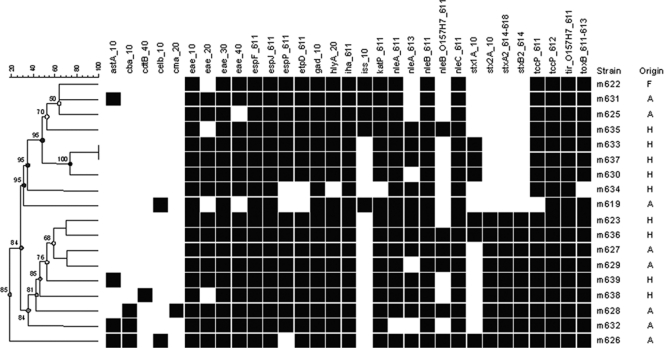
Fig. 2.
Dendrogram based on the Identibac virulence gene microarray results. Only genes that were present in at least one strain have been included. Black boxes indicate genes present. A, animal origin; F, food origin; H, human origin. The survival time is indicated in days. Gene specifics: astA, heat-stable enterotoxin 1; cba, colicin B pore forming; cdtB, cytolethal distending toxin B; celB, endonuclease colicin E2; cma, colicin M (resembles beta-lactam antibiotics); eae, intimin; espF, type III secretion system; espJ, prophage-encoded type III secretion system effector; espP, putative exoprotein precursor; etpD, type II secretion protein; gad, glutamate decarboxylase; hlyA, hemolysin A; iha, adherence protein; iss, increased serum survival; katP, plasmid-encoded catalase peroxidase; nleA, non-LEE-encoded effector A; nleB, non-LEE-encoded effector B; nleC, non-LEE-encoded effector C; stx1A, Shiga toxin 1 A subunit; stx2A, Shiga toxin 2 A subunit; stxA2, Shiga toxin 2 subunit A; stxB2, Shiga toxin 2 subunit B; tccP, Tir-cytoskeleton-coupling protein; tir, translocated intimin receptor; toxB, toxin B potential adhesion.