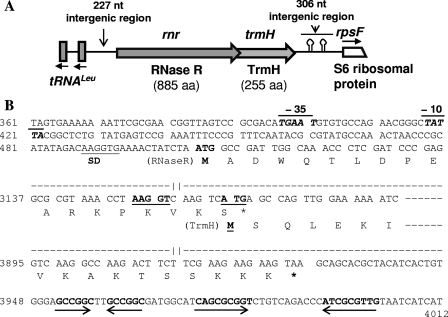
Fig. 1.
Gene organization in the rnr locus of P. syringae Lz4W. (A) Schematic representation of different genes in the 4,468-bp sequenced region (accession no. HQ122447), shown as block arrows on a line. The directions of transcription have been indicated by the arrows. The putative hairpin structures that occur downstream of the rnr-trmH operon and may act as transcription terminator signals have been indicated. Two leucine-tRNA genes transcribed divergently from rnr are shown as vertical blocks. (B) Nucleotide sequence of the relevant regions of the rnr-trmH operon. Only the sequence of the promoter region of the rnr gene, the overlapping region of RNase R and TrmH reading frames, and the downstream region containing the inverted repeats forming putative hairpin structures are shown. The numbers at left of the sequence correspond to the nucleotide numbers in accession no. HQ122447. The sequences not shown here have been indicated by dashed lines with gaps. Marked on the sequences are the “−10” and “−35” regions of the putative rnr promoter shown as a line at the top, putative ribosome binding Shine-Dalgarno (SD) sequences by a line below, and the inverted repeats by opposing arrows below the sequences. Amino acid residues encoded by relevant regions of the reading frames are given in single-letter code. Translational start codons (ATG) are shown in boldface, while the stop codons are indicated by an asterisk (*).