Abstract
A collection of 140 Cryptosporidium parvum isolates previously analyzed by PCR-restriction fragment length polymorphism (PCR-RFLP) and sequence analyses of the small-subunit (SSU) rRNA and 60-kDa glycoprotein (GP60) genes was further characterized by multilocus fragment typing of six minisatellite (MSB and MS5) and microsatellite (ML1, ML2, TP14, and 5B12) loci. Isolates were collected from diarrheic preweaned calves originating from 61 dairy cattle farms in northern Spain. A capillary electrophoresis-based tool combining three different fluorescent tags was used to analyze all six satellites in one capillary. Fragment sizes were adjusted after comparison with sizes obtained by sequence analysis of a selection of isolates for every allele. Size discrepancies at all but the 5B12 locus were found for those isolates that were typed by both techniques, although identical size differences were reported for every allele within each locus. A total of eight alleles were seen at the ML2 marker, which contributed the most to the discriminatory power of the multilocus approach. Multilocus fragment typing clearly improved the discriminatory power of GP60 sequencing, since a total of 59 multilocus subtypes were identified based on the combination of alleles at the six satellite loci, in contrast to the 7 GP60 subtypes previously reported. The majority of farms (38) displayed a unique multilocus subtype, and individual isolates with mixed multilocus subtypes were seen at 22 farms. Bayesian structure analysis based on combined data for both satellite and GP60 loci suggested the presence of two major clusters among the C. parvum isolates from cattle farms in this geographical area.
INTRODUCTION
Cryptosporidium is an obligate intracellular protozoan causing enteric infection in a wide range of mammals, including humans. The genus is composed of multiple genetically distinct but morphologically indistinguishable species and genotypes, with Cryptosporidium parvum being the most common zoonotic species (37). Studies on the prevalence of Cryptosporidium in farm animals have revealed that cattle is the primary nonhuman species impacted by cryptosporidiosis, which is recognized worldwide as one of the most prevalent causes of neonatal diarrhea in domestic livestock (29). A range of host-adapted Cryptosporidium spp. have been described for cattle, but only preweaned calves are frequently infected by C. parvum and contribute significantly to zoonotic cryptosporidiosis (10, 18, 28). Nevertheless, human-specific, animal-specific, and zoonotic C. parvum subtypes have been identified, revealing that molecular subtyping tools are essential for unraveling the transmission dynamics of Cryptosporidium infections in humans and animals, although fundamental questions concerning population genetics, such as the extent of sexual recombination in populations of parasites in the field, are not yet fully resolved (37).
One of the subtyping tools used most commonly is based on sequence analysis of the 60-kDa glycoprotein (GP60) gene, which enables the identification of subtype families within C. parvum as well as several subtypes (alleles) within each family. This locus has the highest resolution as a single marker for subtyping C. parvum isolates, but its use in tracking transmission across regions is limited by the worldwide distribution of the most prevalent subtypes (38). GP60 allelic groups have showed no evidence of segregating in space or differing with respect to geographical diversity, and use of GP60 in a single-locus subtyping method has been reported to underestimate genetic diversity where sexual reproduction occurs (33, 34). In contrast, geographically unique C. parvum subtypes have been identified by multilocus analysis of some minisatellite and microsatellite loci, another class of highly polymorphic markers characterized by allelic variability in repeat length (11). Satellites have been used to investigate the genetic structure and geographic tracking of Cryptosporidium, although the paucity of sequence data for most loci in international databases and the diversity in techniques used to amplify and size alleles have limited comparisons among studies (17).
Satellite polymorphisms have been evaluated by PCR, and allele sizing has been achieved by multilocus sequence typing (5, 6, 9, 12, 13, 24, 35), polyacrylamide gel electrophoresis (11, 31, 32), and automated capillary electrophoresis (CE) with labeled primers, named multilocus fragment typing (2, 7, 15, 19–22, 23, 31, 32). The latter technique has been proven useful for detecting mixed parasite populations in individual samples and as a less expensive alternative to sequencing, although fragments of equal length but with nucleotide substitutions are not differentiated by this method (9, 30, 39).
Most previous CE analyses have relied on the use of a single fluorescent label, which reduces the number of markers that can be analyzed in a single reaction mix. In the current study, a time- and cost-saving capillary electrophoresis-based tool combining three different fluorescent tags was used to analyze six satellites in one capillary. This approach was evaluated to further characterize the genetic polymorphisms exhibited among a group of C. parvum isolates from calves previously analyzed by sequencing of the GP60 gene. We also applied this tool to assess the genetic structure of C. parvum within dairy cattle farms in northern Spain.
MATERIALS AND METHODS
Cryptosporidium isolates.
Genomic DNA samples of C. parvum isolates from 140 diarrheic preweaned calves selected from a previous study examining the Cryptosporidium species and GP60 subtypes responsible for outbreaks of neonatal diarrhea in dairy cattle farms in Spain were included in this analysis (28). These animals originated from 61 farms (mean, 2.29 ± 1.81 calves/farm) in 14 provinces across northern Spain. A single isolate from each of 27 farms and 2 to 9 isolates from each of the remaining 34 farms were included in the molecular analysis. Cryptosporidium species and subtypes in the previous study were determined based on PCR-restriction fragment length polymorphism (PCR-RFLP) and sequence analyses of the small-subunit (SSU) rRNA and GP60 genes, respectively (3, 36).
Capillary electrophoresis.
Each isolate was subtyped at two minisatellite (MSB and MS5) and four microsatellite (ML1, ML2, TP14, and 5B12) loci, using primers described previously or designed by using Primer3 (version 0.4.0 [http://frodo.wi.mit.edu/primer3/]). The mini- and microsatellite fragments were amplified by using simple (MSB, ML2, and 5B12), heminested (MS5 and ML1), and nested (TP14) PCRs under previously described conditions, with some modifications (5–7, 20, 32). The reverse primers used in simple PCRs and the internal reverse primers used in heminested/nested PCRs were 5′ labeled with HEX (4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein), FAM (6-carboxyfluorescein), or TAMRA (6-carboxytetramethylrhodamine), according to the predicted fragment size. The primers used for PCR analysis of all gene targets, the annealing temperatures used, and the sizes of the expected PCR products are listed in Table 1.
Table 1.
Primers used for mini- and microsatellite typing, PCR conditions, and predicted amplicon size ranges
| Locus | GenBank accession no. | Chromosome | Primer | Primer sequence (5′ → 3′) | Repeat motif | Annealing temp (°C) | Fragment size range (bp) | Reference for primer |
|---|---|---|---|---|---|---|---|---|
| ML2 | AF344880 | VI | F | CAATGTAAGTTTACTTATGATTAT | AG | 50 | 180–237 | 6 |
| R | FAM-CGACTATAAAGATGAGAGAAG | 6 | ||||||
| 5B12 | AQ449854 | II | F | TGACGATGAAGATGAGGGAAC | 60 | 134–155 | This study | |
| R | HEX-CAGGACAGATTTAGGAGGAGGA | AT | This study | |||||
| MSB | XM627997 | I | F | GGGAGGCATAGGGATGA | 59 | 32 | ||
| R | TAMRA-CTTTTGATCGCTTCTTTTCCA | AGATAG | 246–324 | 32 | ||||
| F1 | TAATGCCCACCCATCTTCTT | 61 | This study | |||||
| TP14 | XM627041 | VIII | R1 | TCCATCTGGGTCCATTTAGC | CAA | 279–333 | This study | |
| F2 | CTAACGTTCACAGCCAACAGTACC | 62 | 20 | |||||
| R2 | FAM-GTACAGCTCCTGTTCCTGTTG | 20 | ||||||
| F1 | CATGAGCTAAAAATGGTGG | GAG | 55 | 218–242 | 7 | |||
| ML1 | G35348 | III | F2 | CTAAAAATGGTGGAGAATATTC | 50 | 5 | ||
| Ra | HEX-CAACAAAATCTATATCCTC | 5 | ||||||
| F | CTTTCTCATCAACTTGCATG | CCTCCCTCAGCTCCTCCGACTGCA | 60 | 32 | ||||
| MS5 | AQ083744 | VIII | R1 | TTTAGCAGGTGTGGAGCTGA | 192–324 | This study | ||
| R2 | TAMRA-AGCGAGTGGAGGAGCT | 56 | 32 |
The external reverse primer was unlabeled.
PCR mixtures for simple PCRs consisted of 1× PCR buffer, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 1 μM (each) forward and reverse primers, 1 U of Taq polymerase, and 2 μl of the DNA template in a total reaction volume of 20 μl. Templates were subjected to 40 cycles consisting of 94°C for 30 s, the specified annealing temperature for 30 s, and 72°C for 1 min, with an initial denaturation step at 94°C for 5 min and a final extension step at 72°C for 7 min. PCR mixtures for heminested and nested PCRs consisted of 2 μl of DNA template (for primary PCRs) or 2 μl of primary PCR product (for secondary PCRs), 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of each dNTP, a 0.5 μM concentration of each primer, and 1 U of Taq polymerase in a total reaction volume of 20 μl. Each PCR mix for heminested and nested reactions was subjected to 35 cycles consisting of 95°C for 50 s, the marker-specific annealing temperature for 50 s, and 72°C for 1 min, with an initial denaturation step at 95°C for 3 min and a final extension step at 72°C for 10 min.
PCR products were first separated by electrophoresis in 1.5% agarose gels and visualized by staining with GelRed nucleic acid gel stain (Biotium, Hayward, CA) to confirm DNA amplification. According to the amplicon intensity, 0.5- to 2-μl samples of the mini- and microsatellite-labeled PCR products for each C. parvum isolate were then mixed and subjected to capillary electrophoresis on a MegaBACE 500 analyzer (Amersham Biosciences). Dye-labeled amplicons were identified in the form of multicolored electropherograms and were sized automatically using the Et400-R size standard (GE Healthcare) and the aid of Fragment Profiler software (version 1.2). Since C. parvum is a haploid organism and the mini- and microsatellites correspond to single-copy loci, the presence of two separate peaks for a specific locus differing by multiples of the repeat unit was assumed to indicate a mixed infection. In order to confirm the ability of the method to identify mixed parasite populations in individual samples, the labeled PCR products from different alleles for each locus were mixed and subsequently analyzed by fragment analysis as described above. The allele composition of these deliberately mixed samples was as expected.
DNA sequence analysis.
A selection of isolates for each locus was sequenced to confirm size differences and establish identity with alleles determined by CE analysis. For this purpose, at least two representative isolates for each allele were amplified using unlabeled primers and the above-mentioned PCR mixture and cycling conditions. PCR products were purified and sequenced in both directions on a MegaBACE 500 sequencer according to the manufacturer's instructions. Alignment of the consensus sequences obtained and of sequences from the GenBank database was done using Clustal W and edited with BioEdit, version 7.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Allele nomenclature was based on the fragment size (in base pairs) adjusted after comparison with sequence analysis of these representative isolates.
Multilocus analysis.
The multilocus subtype for each isolate was determined based on the combination of alleles at the six loci, and each multilocus subtype was then designated a number. Only isolates that were amplified at all six loci were included in the analysis. When two alleles were identified at one locus, the two potential multilocus subtypes were considered. When more than one locus showed several alleles, multilocus subtypes could not be determined and the isolate was excluded from the population genetic analyses. A factorial correspondence analysis (FCA) was performed by using Genetix software (K. Belkhir, P. Borsa, L. Chikhi, N. Raufaste, and F. Bonhomme, Genetix 4.05 [http://www.genetix.univ-montp2.fr/genetix/genetix.htm]) according to the instructions of the authors. This test places the isolates in a three-dimensional space according to the similarity of their allelic states. The FCA study was based on information from both mini- and microsatellite markers and the previously reported GP60 subtypes. For this purpose, a code for the GP60 subtypes was applied so that the information could be included in the vectorial study. Genetix software was also used to calculate the fixation index (Fst) values, as a measure of the proportion of genetic diversity due to allele frequency differences, for the different origins of the samples. Bayesian genetic structure analysis was carried out by means of Structure software and also included data from all loci (27). This algorithm identifies genetically distinct populations on the basis of allele frequencies. Genetic clusters are constructed from the subtypes identified, with estimation for each isolate of the fraction of its subtype that belongs to each cluster. The conditions for the use of Structure included a burn-in period of 10,000 iterations, 100,000 Markov chain Monte Carlo repetitions, and 3 iterations of each calculation to evaluate variability. An unweighted-pair group method using average linkages (UPGMA) based on the method of Nei (23a) was completed by means of TFPGA (Tools for Population Genetics Analysis) software (M. P. Miller [http://www.marksgeneticsoftware.net/tfpga.htm]).
Nucleotide sequence accession numbers.
Nucleotide sequences generated in this study were deposited in the GenBank database under accession numbers JF342559 to JF342572.
RESULTS
Allele sizing.
PCR products for each isolate showed six colored peaks of fluorescence after analysis by Fragment Profiler software. Each peak corresponded to the length of the PCR product and was allocated to an allele according to the fluorescent label and predicted amplicon size for each marker. Stutter bands differing by 2 bp due to strand slippage of Taq polymerase on microsatellite sequences were sometimes present at both the 5B12 and ML2 loci, but their fluorescence intensity was always much lower than that of the main peak. Allele sizes determined by automated fragment analysis were corrected by comparison with direct sequence analysis of representative isolates. All of these isolates were sequenced successfully, except for alleles longer than 227 bp at the ML2 locus, which had underlying signals in the electropherogram that prevented the accurate readout of sequences. Similarly, repeated attempts to sequence alleles of 215 bp and 191 bp at the MS5 locus were unsuccessful. The correction factors calculated for other alleles of the ML2 and MS5 loci were applied to these unsequenced alleles. Allele sizing was 100% identical for the 5B12 locus for those isolates that were typed by both techniques, but no concordant results were obtained for the remaining five loci, with the fragment analysis providing longer lengths than sequencing. Differences were related directly to locus size and increased from 2 bp at the ML2 locus to 9 bp at the TP14 locus. Nevertheless, it is worth mentioning that size differences were consistent across the entire size range within each locus.
Allele frequencies.
A total of 137 C. parvum isolates from 61 farms were typeable at all six loci. A complete list of the alleles identified for each locus is given in Table 2. The ML2 locus was the most polymorphic marker, with a total of eight alleles of sizes ranging from 185 to 237 bp. Nevertheless, more than 75% of isolates were assigned to only three alleles, namely, ML2-231, ML2-233, and ML2-227. Mixed infections were identified for three samples, all of them harboring the uncommon allele ML2-191. Sequence analysis of selected isolates revealed that ML2-191 was identical to the C. parvum sequence under GenBank accession number AJ308566 (6). Likewise, the previously undescribed allele ML2-185 differed by the contraction of one AG repeat from the reference allele ML2-187 in GenBank (accession number AJ308567) (6).
Table 2.
Allele sizes and number allocation for each of the six mini- and microsatellite loci in C. parvum isolates from diarrheic preweaned calves in northern Spain
| Locus and allele size (bp) (allele no.) | No. (%) of isolates (n = 137) | No. of farms (n = 61) |
|---|---|---|
| ML1 | ||
| 226 (1) | 31 (22.6) | 19 |
| 238 (2) | 98 (71.5) | 43 |
| 226 + 238 | 8 (5.9) | 7 |
| ML2 | ||
| 185 (1) | 1 (0.7) | 1 |
| 191 (2) | 4 (2.9) | 1 |
| 227 (3) | 24 (17.5) | 14 |
| 229 (4) | 11 (8) | 8 |
| 231 (5) | 52 (38) | 22 |
| 233 (6) | 27 (19.8) | 15 |
| 235 (7) | 11 (8) | 8 |
| 237 (8) | 4 (2.9) | 3 |
| 191 + 231 | 1 (0.7) | 1 |
| 191 + 233 | 2 (1.5) | 1 |
| TP14 | ||
| 324 (1) | 37 (27) | 19 |
| 333 (2) | 64 (46.7) | 36 |
| 342 (3) | 20 (14.6) | 12 |
| 333 + 324 | 7 (5.1) | 5 |
| 342 + 324 | 2 (1.5) | 2 |
| 342 + 333 | 6 (4.3) | 5 |
| 342 + 324 + 333 | 1 (0.7) | 1 |
| MS5 | ||
| 215 (1) | 1 (0.7) | 1 |
| 239 (2) | 134 (97.8) | 59 |
| 239 + 191 (3) | 2 (1.5) | 1 |
| 5B12 | ||
| 165 (1) | 44 (32.1) | 19 |
| 167 (2) | 1 (0.7) | 1 |
| 169 (3) | 75 (54.7) | 42 |
| 171 (4) | 12 (8.7) | 6 |
| 165 + 169 | 5 (3.7) | 4 |
| MSB | ||
| 304 (1) | 1 (0.7) | 1 |
| 316 (2) | 3 (2.2) | 2 |
| 322 (3) | 121 (88.3) | 56 |
| 328 (4) | 5 (3.7) | 2 |
| 304 + 322 | 1 (0.7) | 1 |
| 316 + 322 | 1 (0.7) | 1 |
| 310 + 322 (5) | 2 (1.5) | 1 |
| 328 + 322 | 3 (2.2) | 3 |
Four alleles were identified at the 5B12 locus, with most isolates and farms displaying the 5B12-165 and 5B12-169 alleles. All isolates sequenced at this locus were identical to each other in the nonrepeat region and differed by the number of TA repeats in the microsatellite region. The single isolate identified as having the 5B12-167 allele was homologous to the C. parvum sequence under GenBank accession number AQ449854 (11). Three alleles were observed for the TP14 marker, among which TP14-333 was identified on over half of the farms. The presence of mixed infections was common with this marker, and one isolate showed a triallelic pattern. Sequencing of selected isolates revealed that the TP14-324 allele matched the C. parvum reference sequence XM627041 (1) and differed by contractions of the microsatellite repeats from the novel C. parvum alleles TP14-333 and TP14-342. The ML1 microsatellite was the least polymorphic marker, with only two alleles identified in the current study. Individual isolates with mixed infections were also common at this locus. Isolates selected for sequencing were identical to the C1 (ML1-238) and C2 (ML1-226) alleles deposited in GenBank (accession numbers AJ249582 and AJ249583, respectively) (5).
For minisatellite markers, poor genetic diversity was observed for the MS5 locus, since all but one isolate showed the MS5-239 allele. Isolates sequenced at this allele demonstrated 100% identity to the C. parvum reference sequence under GenBank accession number XM625525 (1). It was not possible to directly sequence the MS5-215 and MS5-191 alleles, but their sizes fit the presence of contractions of the minisatellite region (24 bp), and differences in gel bands were clearly visible. A more extensive genetic variability was seen at the MSB locus, although most isolates (88.3%) and farms (56/61 farms) displayed the MSB-322 allele, and four additional alleles (MSB-304, MSB-310, MSB-316, and MSB-328) were restricted to 1 to 3 farms. Isolates sequenced at this allele revealed that MSB-322 was identical to the C. parvum reference sequence under GenBank accession number XM627997 (1) and differed from the remaining alleles by expansions/contractions of the minisatellite region, except for a single isolate identified as MSB-304, which showed two A-to-T transversions downstream of the repeat region.
Multilocus subtypes.
A total of 59 multilocus subtypes were identified based on the combination of alleles at the six mini- and microsatellite loci (Table 3). Some isolates from 17 farms had a biallelic (21 isolates) or triallelic (1 isolate) profile at one locus and were scored as having two or three multilocus subtypes, respectively. Six isolates from 5 farms showed multiple alleles at more than one locus and were excluded from the genetic analyses. Most farms (38) had a distinct multilocus subtype, and a single multilocus subtype was identified for 13 of 32 farms where two or more isolates were collected. Multilocus subtypes were compared to the previously reported subtypes determined by GP60 sequencing and revealed that most of them (48 subtypes) corresponded to a single GP60 allele. Each of the remaining 11 multilocus subtypes corresponded to 2 to 4 different GP60 alleles, resulting in the addition of 14 new combinations.
Table 3.
Multilocus subtypes of Cryptosporidium isolates from dairy cattle farms, based on the combination of alleles at six minisatellite and microsatellite loci and comparison with previously reported GP60 subtypesa
| Multilocus subtype | Allele at locus |
No. of isolates (n = 131)b | No. of farms (n = 58)c | GP60 subtype | |||||
|---|---|---|---|---|---|---|---|---|---|
| ML1 | MS5 | TP14 | MSB | 5B12 | ML2 | ||||
| 1 | 1 | 1 | 2 | 3 | 3 | 4 | 1 | 1 | IIaA15G2 R1 |
| 2 | 1 | 2 | 1 | 1 | 2 | 3 | 1 | 1 | IIdA23G1 |
| 3 | 1 | 2 | 1 | 3 | 1 | 3 | 1 | 1 | IIaA15G2 R1 |
| 4 | 1 | 2 | 1 | 3 | 1 | 5 | 4 | 1 | IIaA15G2 R1 |
| IIaA17G2 R1 | |||||||||
| 5 | 1 | 2 | 1 | 3 | 1 | 7 | 1 | 1 | IIaA15G2 R1 |
| 6 | 1 | 2 | 1 | 3 | 3 | 5 | 7 | 4 | IIaA15G2 R1 |
| IIaA17G2 R1 | |||||||||
| IIdA23G1 | |||||||||
| 7 | 1 | 2 | 1 | 3 | 3 | 6 | 2 | 1 | IIaA16G3 R1 |
| 8 | 1 | 2 | 2 | 3 | 1 | 6 | 1 | 1 | IIaA15G2 R1 |
| 9 | 1 | 2 | 2 | 3 | 3 | 3 | 3 | 2 | IIaA15G2 R1 |
| 10 | 1 | 2 | 2 | 3 | 3 | 4 | 1 | 1 | IIaA15G2 R1 |
| 11 | 1 | 2 | 2 | 3 | 3 | 5 | 2 | 2 | IIaA15G2 R1 |
| 12 | 1 | 2 | 2 | 3 | 3 | 7 | 3 | 3 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| 13 | 1 | 2 | 3 | 3 | 1 | 5 | 1 | 1 | IIaA15G2 R1 |
| 14 | 1 | 2 | 3 | 3 | 3 | 4 | 1 | 1 | IIaA15G2 R1 |
| 15 | 1 | 2 | 3 | 3 | 3 | 5 | 6 | 3 | IIaA15G2 R1 |
| IIaA18G3 R1 | |||||||||
| 16 | 1 | 2 | 3 | 3 | 3 | 6 | 3 | 2 | IIaA15G2 R1 |
| 17 | 1 | 2 | 3 | 3 | 3 | 7 | 1 | 1 | IIaA16G3 R1 |
| 18 | 2 | 2 | 1 | 1 | 1 | 6 | 1 | 1 | IIaA15G2 R1 |
| 19 | 2 | 2 | 1 | 2 | 3 | 6 | 1 | 1 | IIaA15G2 R1 |
| 20 | 2 | 2 | 1 | 3 | 1 | 3 | 2 | 2 | IIaA15G2 R1 |
| 21 | 2 | 2 | 1 | 3 | 1 | 5 | 3 | 2 | IIaA15G2 R1 |
| IIaA17G2 R1 | |||||||||
| 22 | 2 | 2 | 1 | 3 | 1 | 6 | 2 | 1 | IIaA15G2 R1 |
| 23 | 2 | 2 | 1 | 3 | 1 | 7 | 1 | 1 | IIaA15G2 R1 |
| 24 | 2 | 2 | 1 | 3 | 3 | 3 | 1 | 1 | IIaA15G2 R1 |
| 25 | 2 | 2 | 1 | 3 | 3 | 4 | 1 | 1 | IIaA15G2 R1 |
| 26 | 2 | 2 | 1 | 3 | 3 | 5 | 5 | 2 | IIaA15G2 R1 |
| 27 | 2 | 2 | 1 | 3 | 3 | 6 | 8 | 5 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| 28 | 2 | 2 | 1 | 3 | 3 | 7 | 2 | 2 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| 29 | 2 | 2 | 1 | 3 | 3 | 8 | 2 | 1 | IIaA15G2 R1 |
| 30 | 2 | 2 | 1 | 3 | 4 | 4 | 1 | 1 | IIaA16G2 R1 |
| 31 | 2 | 2 | 1 | 3 | 4 | 8 | 1 | 1 | IIaA16G3 R1 |
| 32 | 2 | 2 | 1 | 4 | 3 | 6 | 1 | 1 | IIaA15G2 R1 |
| 33 | 2 | 2 | 2 | 2 | 3 | 5 | 3 | 2 | IIaA15G2 R1 |
| 34 | 2 | 2 | 2 | 3 | 1 | 3 | 10 | 3 | IIaA15G2 R1 |
| 35 | 2 | 2 | 2 | 3 | 1 | 4 | 2 | 1 | IIaA15G2 R1 |
| 36 | 2 | 2 | 2 | 3 | 1 | 5 | 10 | 4 | IIaA15G2 R1 |
| 37 | 2 | 2 | 2 | 3 | 1 | 6 | 6 | 4 | IIaA15G2 R1 |
| 38 | 2 | 2 | 2 | 3 | 1 | 7 | 1 | 1 | IIaA15G2 R1 |
| 39 | 2 | 2 | 2 | 3 | 3 | 1 | 1 | 1 | IIaA15G2 R1 |
| 40 | 2 | 2 | 2 | 3 | 3 | 3 | 2 | 2 | IIaA15G2 R1 |
| 41 | 2 | 2 | 2 | 3 | 3 | 4 | 1 | 1 | IIaA15G2 R1 |
| 42 | 2 | 2 | 2 | 3 | 3 | 5 | 9 | 7 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| 43 | 2 | 2 | 2 | 3 | 3 | 6 | 6 | 3 | IIaA15G2 R1 |
| IIaA17G2 R1 | |||||||||
| 44 | 2 | 2 | 2 | 3 | 3 | 7 | 1 | 1 | IIaA16G3 R1 |
| 45 | 2 | 2 | 2 | 3 | 3 | 8 | 1 | 1 | IIaA15G2 R1 |
| 46 | 2 | 2 | 2 | 3 | 4 | 3 | 2 | 1 | IIaA15G2 R1 |
| 47 | 2 | 2 | 2 | 3 | 4 | 4 | 3 | 2 | IIaA16G2 R1 |
| 48 | 2 | 2 | 2 | 4 | 1 | 3 | 1 | 1 | IIaA15G2 R1 |
| 49 | 2 | 2 | 2 | 4 | 4 | 2 | 4 | 1 | IIaA18G3 R1 |
| 50 | 2 | 2 | 2 | 4 | 4 | 3 | 1 | 1 | IIaA15G2 R1 |
| 51 | 2 | 2 | 2 | 4 | 4 | 4 | 1 | 1 | IIaA16G2 R1 |
| 52 | 2 | 2 | 3 | 3 | 1 | 2 | 1 | 1 | IIaA16G3 R1 |
| 53 | 2 | 2 | 3 | 3 | 1 | 3 | 1 | 1 | IIaA15G2 R1 |
| 54 | 2 | 2 | 3 | 3 | 1 | 5 | 2 | 2 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| 55 | 2 | 2 | 3 | 3 | 1 | 7 | 1 | 1 | IIaA15G2 R1 |
| 56 | 2 | 2 | 3 | 3 | 3 | 1 | 1 | 1 | IIaA15G2 R1 |
| 57 | 2 | 2 | 3 | 3 | 3 | 5 | 9 | 5 | IIaA15G2 R1 |
| IIaA16G3 R1 | |||||||||
| IIaA18G3 R1 | |||||||||
| IIaA19G3 R1 | |||||||||
| 58 | 2 | 2 | 3 | 3 | 3 | 6 | 1 | 1 | IIaA15G2 R1 |
| 59 | 2 | 2 | 3 | 3 | 3 | 7 | 2 | 1 | IIaA15G2 R1 |
The number allocation for alleles is indicated in Table 2.
Samples with mixed infections at one locus were allocated to the corresponding multilocus subtype.
Three farms gave only samples with mixed infections at more than one locus and were excluded from the genetic analyses.
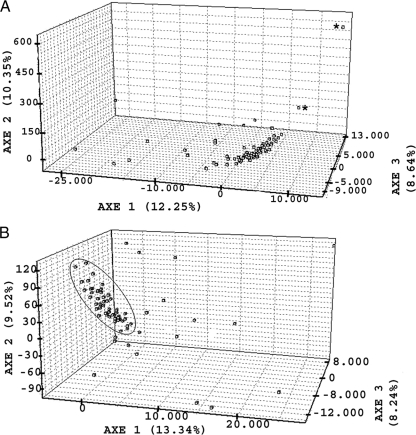
The UPGMA analysis did not provide any relevant information, since the validity of the branches was not supported by bootstrap values. In contrast, results of the FCA study showed that the discriminatory power was clearly improved by the additional information obtained from satellite typing. The only two isolates showing the GP60 subtype within family IId (IIdA23G1) were readily differentiated from each other and clearly separated from the remaining isolates (Fig. 1A). A more detailed three-dimensional distribution was seen when the former were excluded from the vectorial study, with most isolates forming a closely related group and fewer isolates unevenly dispersed in the three-dimensional scatterplot (Fig. 1B). Results of the Bayesian structure analysis are summarized in Fig. 2. The maximum likelihood was obtained for a number of clusters, with the most probable number of clusters (K) being 2, represented by different colors in the figure. An isolate was considered to belong to one of the clusters only if the probability of it belonging was higher than 0.8. Otherwise, the isolate was considered to have a mixed origin. Altogether, 54% of the samples belonged to one of the clusters, while the remaining isolates were considered to have a mixed origin. The estimated Fst value was 0.6088. This value is highly significant, since random permutation tests based on up to 10,000 reiterations provided 0.00% values superior to this one. This indicates the existence of significant genetic differences among some of the sampling origins. In fact, the pairwise Fst analysis (data not shown) provided a very wide range of values (0 to 1) for this parameter in the different pairs of sampling origins.
Fig. 1.
FCA study of Cryptosporidium parvum isolates from diarrheic preweaned calves in northern Spain. (A) All 131 C. parvum isolates were analyzed, including the only 2 isolates showing the subtype IIdA23G1 (*). (B) Vectorial analysis when the latter isolates were excluded.
Fig. 2.
Bayesian genetic structure analysis of 131 C. parvum isolates from 58 dairy cattle farms, as inferred by Structure software and based on total multilocus information, including information on 6 mini- and microsatellite markers and GP60 subtypes. The bar plot shows the most probable number of clusters (K = 2). Each isolate is represented by a single vertical line divided into two gray tonalities. Each tonality represents one cluster, and the length of the gray segment shows the isolate's estimated proportion of membership in that population.
DISCUSSION
The results of the current study reveal that CE analysis is a sensitive and discriminatory approach for subtyping Cryptosporidium isolates. Peaks were well defined even for those samples with bands scarcely visible on gel electrophoresis, and the resolution of electrophoresis in the automated analyzer was so high that we could easily separate peaks differing by only 2 bp, corresponding to a dinucleotide repeat. Moreover, the tool was useful for detecting mixed infections in individual samples, a finding which is consistent with other studies and the fact that natural, uncloned parasite populations were analyzed (13, 20, 21, 25, 31). A potential limitation of CE tools relates to the fact that they do not yield DNA sequence data, which substantially limits comparisons among studies (17). On the other hand, a lack of agreement between the actual allele size, which can be confirmed only by sequencing, and sizes obtained by automated CE has been described. This result seems independent of the fluorescent dye but might be due to different migration patterns in the capillary of the size standard and the satellite loci, which impairs the comparison of data collected across laboratories on different instruments (8, 26).
Analyses of PCR-labeled markers have been applied for typing of Cryptosporidium isolates from human and livestock origins, but few studies have calibrated the sizes of satellite fragments by comparison with reference sequenced material. Some studies have reported concordant results (2, 19–21), while a recent analysis of C. parvum isolates from domestic ruminants concluded that CE provides fragment sizes that are 4 bp longer than those obtained by sequencing or high-resolution gel electrophoresis for both the ML1 and ML2 microsatellites (P. Díaz et al., unpublished data). In the current study, size discrepancies between fragment analysis and sequencing were found at all but one locus, although identical size differences were reported for every allele within each locus and enabled us to establish a unique correction factor for each marker. These observations explain the lack of identity between some alleles described by different authors and highlight the need to carry out an in-house calibration by sequencing to assign sizes to the fragments of interest. It is also worth mentioning that all alleles differed by multiples of the repeat unit, suggesting that polymorphisms were due to changes in the numbers of satellite units rather than to insertions or deletions of nucleotides within the fragment, although single-nucleotide mutations outside the repeat region were occasionally seen in some sequenced isolates.
The previous characterization by sequence analysis of the GP60 gene revealed that all isolates belonged to GP60 subtypes previously described for human patients (28). This observation was confirmed by polymorphisms at some microsatellite loci and further supports the hypothesis that most isolates from diarrheic calves in northern Spain have zoonotic potential. The ML2 locus was the most variable marker and contributed considerably to the discriminatory power of the multilocus approach, since its exclusion dramatically reduced the number of multilocus subtypes to 22, while a much more modest genetic diversity was seen at the ML1 locus, with only two alleles identified among the cattle population. All alleles found at both the ML1 and ML2 loci have also been identified in humans and livestock in Italy, The Netherlands, Denmark, Australia, and the United Kingdom, except for the novel allele ML2-185, seen in a single isolate in this study (5–7, 9, 14–16, 19, 21, 35). As expected for microsatellites consisting of long repeats of dinucleotide units, stutter bands were seen, especially for the longest alleles at the ML2 locus (26). These artifacts did not hinder allocation to a specific allele in an automated sequence analyzer, as opposed to the case described by Alves et al. (2), but prevented accurate sequence analysis, as reported by Díaz et al. (unpublished data).
Comparison between alleles identified at the remaining loci and C. parvum alleles reported by other authors could not be done because of either the different methods used for sizing of PCR fragments or the use of different primer sets. No similarities were seen for alleles at the 5B12 locus, whose genetic diversity seems to have strong geographical variations. It was recorded as the most variable of 14 microsatellites by Feng et al. (11), while no polymorphisms were seen at this locus in C. parvum isolates from cattle farms in Israel (31). In our study, the 5B12 locus was the second most polymorphic microsatellite, but its contribution to the discriminatory power of the multilocus analysis was reduced by the fact that more than 90% of the isolates were allocated to only two alleles. In contrast, the three alleles identified at the TP14 locus were more evenly distributed, with remarkably large numbers of samples consisting of mixed infections reported by this marker. The most prevalent allele, TP14-333, could correspond to a fragment of 331 bp identified in all C. parvum isolates from calves in Sweden (4). Likewise, our TP14-324 allele could be homologous to an allele of 322 bp found in isolates from humans in the same country, suggesting a zoonotic potential (22).
Both minisatellite loci provided poor resolution. Five alleles were seen at the MSB locus, although all but nine isolates showed the MSB-322 allele in either single or mixed infections, which could correspond to a fragment of 324 bp reported as the most prevalent allele at this locus for cattle farms in Turkey and Israel (31). Similarly, most of the C. parvum isolates were indistinguishable at the MS5 locus, having the MS5-239 allele. This lack of diversity is similar to that reported by Leoni et al. (19), who found six different alleles among C. parvum isolates from humans and livestock in the United Kingdom, although most of them (93%) shared an allele of 328 bp, including all 17 isolates from calves and lambs. Similarly, this minisatellite was monomorphic among C. parvum isolates from calves and goats in Israel, Turkey, and France (25, 31). In contrast, the MS5 locus was described as one of the most polymorphic markers among C. parvum isolates from humans and livestock in Scotland, with a total of 13 different alleles (21).
The combined results of fragment size analysis of all six markers provided a more robust analysis than that with the previous typing based on sequencing of the GP60 gene. A total of 59 different multilocus subtypes were identified, and the majority of farms displayed a unique multilocus fingerprint. The high genetic diversity seen among farms with similar methods of husbandry in a limited geographical area demonstrates that multilocus satellite analysis is a discriminatory approach for typing C. parvum. The finding of individual isolates with mixed multilocus subtypes on more than one-third of the farms (22/61 farms) also reveals that infections by genetically distinct subpopulations of C. parvum are common in calves and supports the complexity of cryptosporidial transmission on ruminant farms. These observations were supported by a recent multilocus fragment analysis in northeastern Spain (J. Quílez et al., unpublished data). That study concluded that alleles that are unreported or scarcely seen in calves, such as ML2-191, ML2-193, MSB-304, and MSB-310, are the most prevalent in diarrheic lambs and goat kids, which highlights the value of these markers in the epidemiological investigation of outbreaks.
Bayesian structure analysis based on the combined data for both the satellite and GP60 markers suggested the presence of two major clusters including approximately half of the isolates, while the remaining isolates showed a mixed origin. This finding indicates the existence of a genetic structure among the C. parvum isolates from cattle farms in this geographical area and is consistent with a previous analysis revealing the emergence of genetically distinct C. parvum populations on different farms (31). Other studies have also revealed that satellites are useful markers for differentiating genetic populations within Cryptosporidium species. Mallon et al. (20) defined 38 multilocus subtypes among a collection of C. hominis and C. parvum isolates from both humans and cattle by using three minisatellite and four microsatellite markers and found evidence of four genetically isolated populations, including one group of human-specific isolates comprised of two closely related multilocus types (clonal isolates), while a second group infecting humans and animals showed a panmictic population structure. Hunter et al. (15) also reported the presence of two clones of human-adapted strains of C. parvum, based on polymorphisms of three microsatellite loci, as well as a third cluster containing strains from persons reporting contact with animals.
In conclusion, the multilocus fragment analysis used in the study is a sensitive and cost-effective approach which detects extensive polymorphisms in C. parvum and provides us with a powerful tool for strain typing and epidemiological tracking. Most of the loci used are useful in distinguishing C. parvum subtypes, although a standardized protocol for amplifying and sizing fragments is needed to investigate the significance of the diverse multilocus subtypes within C. parvum in relation to geographical factors.
ACKNOWLEDGMENTS
This work was supported by funds from Spanish (AGL2009-10590) and regional (DGA-B82) research programs.
We thank Teresa Tejedor (Faculty of Veterinary Sciences, University of Zaragoza) and J. Luis Juaristi (Rancho Las Nieves) for their helpful assistance. We also thank Rachel M. Chalmers and Stephen J. Hadfield (UK Cryptosporidium Reference Unit) for a thoughtful and careful review of the manuscript.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Abrahamsen M. S., et al. 2004. Complete genome sequence of the Apicomplexan, Cryptosporidium parvum. Science 304:441–445 [DOI] [PubMed] [Google Scholar]
- 2. Alves M., Matos O., Antunes F. 2003. Microsatellite analysis of Cryptosporidium hominis and Cryptosporidium parvum in Portugal: a preliminary study. J. Eukaryot. Microbiol. 50:529–530 [DOI] [PubMed] [Google Scholar]
- 3. Alves M., et al. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Björkman C., Mattsson J. G. 2006. Persistent infection in a dairy herd with an unusual genotype of bovine Cryptosporidium parvum. FEMS Microbiol. Lett. 254:71–74 [DOI] [PubMed] [Google Scholar]
- 5. Cacciò S., et al. 2000. A microsatellite marker reveals population heterogeneity within humans and animal genotypes of Cryptosporidium parvum. Parasitology 120:237–244 [DOI] [PubMed] [Google Scholar]
- 6. Cacciò S., Spano F., Pozio F. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082–1086 [DOI] [PubMed] [Google Scholar]
- 7. Chalmers R. M., et al. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397–410 [DOI] [PubMed] [Google Scholar]
- 8. Delmotte F., Leterme N., Simón J. C. 2001. Microsatellite allele sizing: difference between automated capillary electrophoresis and manual technique. Biotechniques 31:814–818 [PubMed] [Google Scholar]
- 9. Enemark H. L., et al. 2002. Molecular characterization of Danish Cryptosporidium parvum isolates. Parasitology 125:331–341 [DOI] [PubMed] [Google Scholar]
- 10. Fayer R., Santín M., Trout J. M. 2007. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet. Parasitol. 145:260–266 [DOI] [PubMed] [Google Scholar]
- 11. Feng X., et al. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gatei W., et al. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53(Suppl. 1):S43–S48 [DOI] [PubMed] [Google Scholar]
- 13. Gatei W., et al. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197–205 [DOI] [PubMed] [Google Scholar]
- 14. Huetink R. E., van der Giessen J. W., Noordhuizen J. P., Ploeger H. W. 2001. Epidemiology of Cryptosporidium spp. and Giardia duodenalis on a dairy farm. Vet. Parasitol. 102:53–67 [DOI] [PubMed] [Google Scholar]
- 15. Hunter P. R., et al. 2007. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg. Infect. Dis. 13:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunter P. R., et al. 2008. Microsatellite typing of Cryptosporidium parvum in isolates from a waterborne outbreak. J. Clin. Microbiol. 46:3866–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jex A. R., Smith H. V., Monis P. T., Campbell B. E., Gasser R. B. 2008. Cryptosporidium—biotechnological advances in the detection, diagnosis and analysis of genetic variation. Biotechnol. Adv. 26:304–317 [DOI] [PubMed] [Google Scholar]
- 18. Langkjær R. B., Vigre H., Enemark H. L., Maddox-Hyttel C. 2007. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology 134:339–350 [DOI] [PubMed] [Google Scholar]
- 19. Leoni F., Mallon M. E., Smith H. V., Tait A., McLauchlin J. 2007. Multilocus analysis of Cryptosporidium hominis and Cryptosporidium parvum from sporadic and outbreak-related human cases and C. parvum from sporadic cases in livestock in the United Kingdom. J. Clin. Microbiol. 45:3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mallon M., et al. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407–417 [DOI] [PubMed] [Google Scholar]
- 21. Mallon M., MacLeod A., Wastling J. M., Smith H., Tait A. 2003. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect. Genet. Evol. 3:207–218 [DOI] [PubMed] [Google Scholar]
- 22. Mattsson J. G., Insulander M., Levad M., Björkman C., Svenungsson B. 2008. Molecular typing of Cryptosporidium associated with a diarrhoea outbreak identifies two sources of exposure. Epidemiol. Infect. 136:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morrison L. J., et al. 2008. The population structure of the Cryptosporidium parvum population in Scotland: a complex picture. Infect. Genet. Evol. 8:121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a. Nei M. 1972. Genetic distance between populations. Am. Nat. 106:283–292 [Google Scholar]
- 24. Ng J., MacKenzie B., Ryan U. 2010. Longitudinal multi-locus molecular characterisation of sporadic Australian human clinical cases of cryptosporidiosis from 2005 to 2008. Exp. Parasitol. 125:348–356 [DOI] [PubMed] [Google Scholar]
- 25. Ngouanesavanh T., et al. 2006. Cryptosporidium population genetics: evidence of clonality in isolates from France and Haiti. J. Eukaryot. Microbiol. 53(Suppl. 1):S33–S36 [DOI] [PubMed] [Google Scholar]
- 26. Pasqualotto A. C., Denning D. W., Anderson M. J. 2007. A cautionary tale: lack of consistency in allele sizes between two laboratories for a published multilocus microsatellite typing system. J. Clin. Microbiol. 45:522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quílez J., et al. 2008. Cryptosporidium species and subtype analysis from dairy calves in Spain. Parasitology 135:1613–1620 [DOI] [PubMed] [Google Scholar]
- 29. Santín M., Trout J. M. 2008. Livestock, p. 451–483 In Fayer R., Xiao L. (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 30. Schindler A. R., Abs El-Osta Y. G., Stevens M., Sinclair M. I., Gasser R. B. 2005. Capillary electrophoretic analysis of fragment length polymorphism in ribosomal markers of Cryptosporidium from humans. Mol. Cell. Probes 19:394–399 [DOI] [PubMed] [Google Scholar]
- 31. Tanriverdi S., et al. 2006. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl. Environ. Microbiol. 72:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanriverdi S., Widmer G. 2006. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect. Genet. Evol. 6:113–122 [DOI] [PubMed] [Google Scholar]
- 33. Widmer G. 2009. Meta-analysis of a polymorphic surface glycoprotein of the parasitic protozoa Cryptosporidium parvum and Cryptosporidium hominis. Epidemiol. Infect. 137:1800–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Widmer G., Lee Y. 2010. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl. Environ. Microbiol. 76:6639–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wielinga P. R., et al. 2008. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int. J. Parasitol. 38:809–817 [DOI] [PubMed] [Google Scholar]
- 36. Xiao L., et al. 2001. Molecular characterisation of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao L., Feng Y. 2008. Zoonotic cryptosporidiosis. FEMS Immunol. Med. Microbiol. 52:309–323 [DOI] [PubMed] [Google Scholar]
- 38. Xiao L., Ryan U. 2008. Molecular epidemiology, p. 119–171 In Fayer R., Xiao L. (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 39. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]