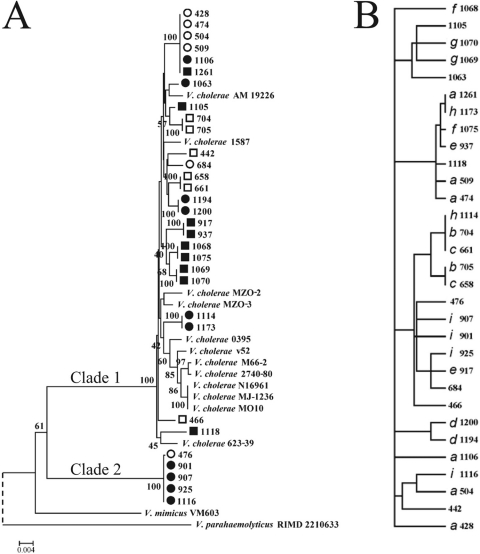
Fig. 1.
Consensus neighbor-joining tree of GBE isolates. (A) A tree was constructed from concatenated sequences of 5 loci, including gapA, gyrB, pyrH, recA, and topA sequences (2,792 bp), using the Jukes-Cantor model of substitution for 31 GBE isolates from Nannie Island (circles) and Oyster River (squares) collected in 2008 (open symbols) and 2009 (filled symbols), as well as five serotype O1 (four El Tor biotypes and one classical biotype) reference strains and eight clinical non-O1 reference strains. V. mimicus VM603 and V. parahaemolyticus RIMD 2210633 are included as tree roots for comparison, but the V. parahaemolyticus branch was shortened in the figure (dashed line). Bootstrap values are from 1,000 replicates. Bar, 0.2% divergence. (B) MLST clonal complexes are labeled with an italic letter to differentiate the clones in the PARS tree, which is based on the discrete levels of the most variable eight phenotypes tested.