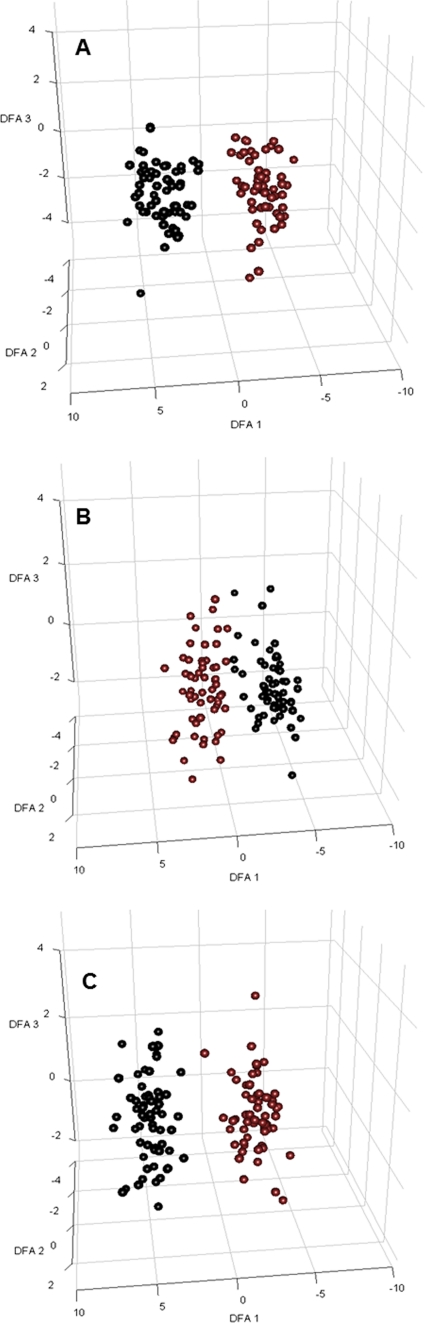
Fig. 3.
DFA for sample visualization of a large data set used for construction of the classification model (controlled contamination). GC-MS metabolite data successfully distinguished samples from contaminated versus noncontaminated microalgal flasks at different time points. Projections of the log-transformed intracellular metabolite data from 120 samples into 3D space show two distinct clusters of the two data classes. For each sample, the projection values were calculated as the linear combination of metabolite values determined by DFA. Only metabolites that were detected in all samples for each data class were used for the analysis. Black, samples from flasks contaminated by P. aeruginosa; red, samples from noncontaminated microalgal flasks. (A) Three hours after contamination; (B) 6 h after contamination; (C) 9 h after contamination.