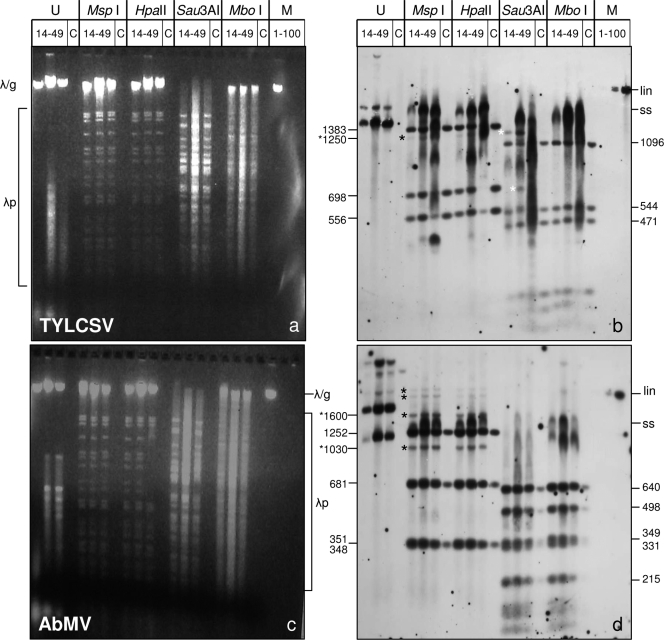
Fig. 1.
Methylation-sensitive restriction enzyme digestion of viral DNA by use of isoschizomers. Total nucleic acids (300 ng DNA [each]) from N. benthamiana plants infected systemically with AbMV or TYLCSV and harvested at 14, 21, or 49 dpi were digested with two sets of isoschizomers: MspI (blocked by cytosine methylation of the external C) with HpaII (blocked by each of the cytosine methylations) (at CCGG) and Sau3AI (blocked by cytosine methylation but not by adenine methylation) with MboI (blocked by cytosine as well as adenine methylation) (at GATC). Untreated samples (U) with the same amounts of DNA were loaded in parallel. The samples were electrophoresed in 1.4% agarose gels (3 h, 120 V), stained with ethidium bromide (a and c), blotted onto nylon membranes, and detected with virus-specific full-length probes for TYLCSV (b) and AbMV A (d). RCA products (1 μl; diluted 1:50) from correspondingly infected plants were digested with the respective enzymes and served as size markers for completely unmethylated DNA fragments (C). Hybridization standards (M) were 1, 10, or 100 pg of linearized full-length dsDNA fragments (lin) of AbMV or TYLCSV. In order to control the completeness of digestion, 300 ng of lambda DNA was supplied to each enzyme reaction mix, forming band patterns (λp) in ethidium bromide-stained gels. Undigested λ DNA was expected to migrate to the same position as genomic plant DNA (λ/g). Geminivirus fragments which were not detected in the controls are marked by black and white asterisks. Expected fragment sizes (in base pairs) of the respective digestion products, calculated from the sequences, are shown to the left and right of the panels. Note that ssDNA tends to smear and to create more diffuse bands than those of dsDNA, as seen after Southern blot hybridization. Bands in undigested samples of ethidium bromide-stained gels refer to the usual genomic DNA and host RNA species (not indicated).