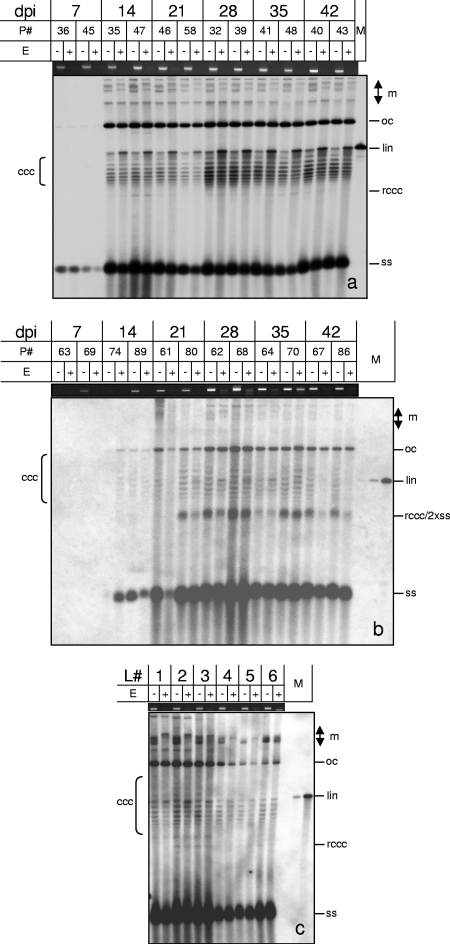
Fig. 5.
Methylation-dependent restriction enzyme digestion of AbMV (a)- or TYLCSV (b)-infected N. benthamiana plants (P#, plant numbers) harvested at the indicated dpi. (c) In addition, individual leaves (L#, starting with the youngest leaf as L1) of ornamental Abutilon plants which were naturally infected with AbMV were collected. Total nucleic acids (60 ng DNA [each]) were digested with McrBC (E+) or not digested (E−), separated electrophoretically in 1.4% agarose gels containing chloroquine (50 μg/ml [a] or 20 μg/ml [b and c]), and analyzed by Southern blot hybridization against the respective probes (AbMV DNA A or TYLSCV lacking each intergenic region). Viral DNA forms are indicated as described in the legend to Fig. 1 and also as multimeric (m), open circular (oc), or relaxed covalently closed circular (rccc) DNA. As a loading control and to ensure complete digestion, equal amounts of every sample were separated in parallel in 1.4% agarose gels and stained with ethidium bromide to show the genomic plant DNA. Hybridization standards (M) were 100 pg (a) or 1, 10, or 100 pg (b and c) of linearized full-length dsDNA fragments (lin) of AbMV DNA A or TYLCSV. For each time point, two of five plants were selected randomly and used for digestion (a and b).