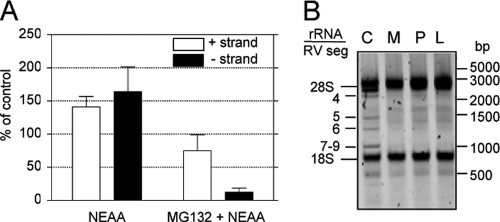
Fig. 4.
Proteasome inhibitors block the replication of the viral genome. (A) The levels of RRV gene 10-negative (closed bars) and -positive (open bars) strands were detected by real-time PCR. Cells were grown in 6-well plates, infected in the presence or absence of 10 μM MG132, and at 9 hpi RNA was purified by TRIzol extraction and the abundance of both strands of gene 10 was evaluated as described in Materials and Methods. Note that the positive strand represents the sum of the mRNA plus the sense strand in the genomic dsRNA, while the negative strand is only present in the genomic segments. The results represent the arithmetic means ± SEM of 3 independent experiments performed in triplicate and are expressed as a percentage of the level of the corresponding (negative- or positive-sense) viral RNA in cells incubated in cell medium free of both MG132 and NEAA. (B) Total RNA was extracted from infected cells incubated with NEAA (C), NEAA plus 10 μM MG132 (M), 20 μM proteasome inhibitor (P), or 7.5 μM lactacystin (L). One microgram of each RNA sample was run in a 1.5% TAE agarose gel, and the RNA was visualized by ethidium bromide staining using a Typhoon Trio scanner. The migration positions of rotavirus dsRNA segments (RV seg) and rRNAs are indicated. One representative experiment of 3 performed is shown.